The Wisconsin Blog
I just wanted to notify anyone who might be interested in following my more personal reflections on my month in Wisconsin (and in Michigan over EDAR4) that I will be updating my Wisconsin Blog at this site (hopefully) regularly. The blog updates are not visible on the first page, so you will have to actively go to the Wisconsin Blog page by clicking in the upper right of the page.
My first day in Madison


Published paper: The Calanus glacialis mitogenome
Mitochondrial DNA Part B today published a mitochondrial genome announcement paper (1) in which I was involved in doing the assemblies and annotating them. The paper describes the mitogenome of Calanus glacialis, a marine planktonic copepod, which is a keystone species in the Arctic Ocean. The mitogenome is 20,674 bp long, and includes 13 protein-coding genes, 2 rRNA genes and 22 tRNA genes. While this is of course note a huge paper, we believe that this new resource will be of interest in understanding the structure and dynamics of C. glacialis populations. The main work in this paper has been carried out by Marvin Choquet at Nord University in Bodø, Norway. So hats off to him for great work, thanks Marvin! The paper can be read here.
Reference
- Choquet M, Alves Monteiro HJ, Bengtsson-Palme J, Hoarau G: The complete mitochondrial genome of the copepod Calanus glacialis. Mitochondrial DNA Part B, 2, 2, 506–507 (2017). doi: 10.1080/23802359.2017.1361357 [Paper link]
New employment – same work
Today, I started my new position at the University of Gothenburg as a non-tenured assistant professor (forskarassistent)*. In essence, this means that I have a position funded by my own grant until the end of 2020, although I will be on a leave-of-absence while doing my PostDoc with Jo Handelsman in Wisconsin. Speaking of which, I will be leaving to the US on Thursday next week for a month of setting things up at her lab (and also going to the EDAR4 conference in Lansing). I will return to Sweden in mid-September and leave for the US for real early next year.
In terms of actual work, this change of position will not mean very much at the moment. I will continue to do the same things for some time, and I will remain closely associated with Joakim Larsson’s lab at the Dept. of Infectious Diseases. And luckily, I will retain my lovely roommates for at least the time being. In the long run, however, this means that I will shift my research focus slightly, away from antibiotic resistance risk management towards interactions in microbial communities (still related to antibiotics though). Exciting times ahead!
Note
* For some reason, the university administration refuses to call this position assistant professor in English at this time, instead referring to the position as “Postdoctoral research fellow”. I guess that it might be bloody annoying explaining that this is not the same as “postdoctoral researcher” and virtually everywhere else would be called “(non-tenured) assistant professor”, but then on the other hand, who cares about titles anyway?
Published paper: Helicobacter pylori vs. the viable gastric microbiota
I have just returned from a week of vacation in Sicily (almost without internet access), so I am a tad late to this news, but earlier this week Infection and Immunity published our paper on the Helicobacter pylori transcriptome in gastric infection (and early stages of carcinogenesis), and how that relates to the transcriptionally active microbiota in the stomach environment (1). This paper has been long in the making (an earlier version of it was included in Kaisa Thorell’s PhD thesis (2)), but some late additional analyses did substantially strengthen our confidence in the suggestions we got from the original data.
In the paper (1) we use metatranscriptomic RNAseq to investigate the composition of the viable microbial community, and at the same time study H. pylori gene expression in stomach biopsies. The biopsies were sampled from individuals with different degrees of H. pylori infection and/or pre-malignant tissue changes. We found that H. pylori completely dominates the microbiota in infected individuals, but (somewhat surprisingly) also in the majority of individuals classified as H. pylori uninfected using traditional methods. This confirms previous reports that have detected minute quantities of H. pylori also in presumably uninfected individuals (3-6), and raises the question of how large part of the human population (if any) that is truly not infected/colonized by H. pylori. The abundance of H. pylori was correlated with the abundance of Campylobacter, Deinococcus, and Sulfurospirillum. It is unclear, however, if these genera only share the same habitat preferences as Helicobacter, or if they are specifically promoted by the presence of H. pylori (or tissue changes induced by it). We also found that genes involved in pH regulation and nickel transport were highly expressed in H. pylori, regardless of disease stage. As far as we know, this study is the first to use metatranscriptomics to study the viable microbiota of the human stomach, and we think that this is a promising approach for future studies on pathogen-microbiota interactions. The paper (in unedited format) can be read here.
References
- Thorell K, Bengtsson-Palme J, Liu OH, Gonzales RVP, Nookaew I, Rabeneck L, Paszat L, Graham DY, Nielsen J, Lundin SB, Sjöling Å: In vivo analysis of the viable microbiota and Helicobacter pylori transcriptome in gastric infection and early stages of carcinogenesis. Infection and Immunity, accepted manuscript (2017). doi: 10.1128/IAI.00031-17 [Paper link]
- Thorell K: Multi-level characterization of host and pathogen in Helicobacter pylori-associated gastric carcinogenesis. Doctoral thesis, Institute of Biomedicine, Sahlgrenska Academy, University of Gothenburg (2014). [Link]
- Bik EM, Eckburg PB, Gill SR, Nelson KE, Purdom EA, Francois F, Perez-Perez G, Blaser MJ, Reman DA: Molecular analysis of the bacterial microbiota in the human stomach. PNAS, 103:732-737 (2006).
- Dicksved J, Lindberg M, Rosenquist M, Enroth H, Jansson JK, Engstrand L: Molecular characterization of the stomach microbiota in patients with gastric cancer and in controls. Journal of Medical Microbiology, 58:509-516 (2009).
- Maldonado-Contreras A, Goldfarb KC, Godoy-Vitorino F, Karaoz U, Contreras M, Blaser MJ, Brodie EL, Dominguez-Bello MG: Structure of the human gastric bacterial community in relation to Helicobacter pylori status. ISME Journal, 5:574-579 (2011).
- Li TH, Qin Y, Sham PC, Lau KS, Chu KM, Leung WK: Alterations in Gastric Microbiota After H. Pylori Eradication and in Different Histological Stages of Gastric Carcinogenesis. Scientific Reports, 7:44935 (2017).
Major ITSx update (beta version)
Today, I am very happy to announce that after years in the making and months in testing, the next generation of ITSx, version 1.1, is ready to step into the public light and scrutiny. I have today uploaded a public beta version of the ITSx 1.1 release, which I encourage everyone that have enjoyed using ITSx to try out.
The 1.1 release of ITSx includes a wide range of new feature, including:
- A 2-10x performance increase (depending on the dataset), since ITSx now utilizes hmmsearch instead of hmmscan to detect the ITS regions and distributes the CPU cores better
- Improved ITS detection among fungi and chlorophyta, by addition of new HMM-profiles
- The HMM profile format for ITSx has been updated to HMMER3/f (thus ITSx now requires HMMER version 3.1 or later)
- Better handling of interrupted HMMER searches
- Added the
--require_anchoroption to only include sequences where the complete anchor is found in the output - Added the possibility for partial sequence output for the SSU, LSU and 5.8S regions
- Fixed a bug causing problems when reading sequence data from standard input
A lot of the code has changed in this version, which means that there might still be bugs lingering in the program. Since I will be on vacation throughout July, I encourage everyone to submit bug reports and questions, but I will not promise to respond to them until in August.
I hope that you will enjoy this new ITSx release, which you can download here. Happy barcoding!
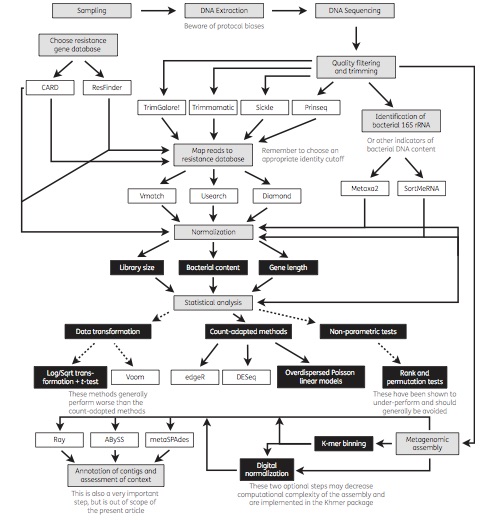
Published paper: Investigating resistomes using metagenomics
Today, a review paper which I wrote together with Joakim Larsson and Erik Kristiansson was published in Journal of Antimicrobial Chemotherapy (1). We have for a long time used metagenomic DNA sequencing to study antibiotic resistance in different environments (2-6), including in the human microbiota (7). Generally, our ultimate purpose has been to assess the risks to human health associated with resistance genes in the environment. However, a multitude of methods exist for metagenomic data analysis, and over the years we have learned that not all methods are suitable for the investigation of resistance genes for this purpose. In our review paper, we describe and discuss current methods for sequence handling, mapping to databases of resistance genes, statistical analysis and metagenomic assembly. We also provide an overview of important considerations related to the analysis of resistance genes, and end by recommending some of the currently used tools, databases and methods that are best equipped to inform research and clinical practice related to antibiotic resistance (see the figure from the paper below). We hope that the paper will be useful to researchers and clinicians interested in using metagenomic sequencing to better understand the resistance genes present in environmental and human-associated microbial communities.
References
- Bengtsson-Palme J, Larsson DGJ, Kristiansson E: Using metagenomics to investigate human and environmental resistomes. Journal of Antimicrobial Chemotherapy, advance access (2017). doi: 10.1093/jac/dkx199 [Paper link]
- Bengtsson-Palme J, Boulund F, Fick J, Kristiansson E, Larsson DGJ: Shotgun metagenomics reveals a wide array of antibiotic resistance genes and mobile elements in a polluted lake in India. Frontiers in Microbiology, 5, 648 (2014). doi: 10.3389/fmicb.2014.00648 [Paper link]
- Lundström S, Östman M, Bengtsson-Palme J, Rutgersson C, Thoudal M, Sircar T, Blanck H, Eriksson KM, Tysklind M, Flach C-F, Larsson DGJ: Minimal selective concentrations of tetracycline in complex aquatic bacterial biofilms. Science of the Total Environment, 553, 587–595 (2016). doi: 10.1016/j.scitotenv.2016.02.103 [Paper link]
- Bengtsson-Palme J, Hammarén R, Pal C, Östman M, Björlenius B, Flach C-F, Kristiansson E, Fick J, Tysklind M, Larsson DGJ: Elucidating selection processes for antibiotic resistance in sewage treatment plants using metagenomics. Science of the Total Environment, 572, 697–712 (2016). doi: 10.1016/j.scitotenv.2016.06.228 [Paper link]
- Pal C, Bengtsson-Palme J, Kristiansson E, Larsson DGJ: The structure and diversity of human, animal and environmental resistomes. Microbiome, 4, 54 (2016). doi: 10.1186/s40168-016-0199-5 [Paper link]
- Flach C-F, Pal C, Svensson CJ, Kristiansson E, Östman M, Bengtsson-Palme J, Tysklind M, Larsson DGJ: Does antifouling paint select for antibiotic resistance? Science of the Total Environment, 590–591, 461–468 (2017). doi: 10.1016/j.scitotenv.2017.01.213 [Paper link]
- Bengtsson-Palme J, Angelin M, Huss M, Kjellqvist S, Kristiansson E, Palmgren H, Larsson DGJ, Johansson A: The human gut microbiome as a transporter of antibiotic resistance genes between continents. Antimicrobial Agents and Chemotherapy, 59, 10, 6551–6560 (2015). doi: 10.1128/AAC.00933-15 [Paper link]
Talk on emission limits in Stockholm
In two weeks time, on the 15th of June, I will participate in a seminar organised by Landstingens nätverk för läkemedel och miljö (the Swedish county council network for pharmaceuticals and environment; the seminar will be held in Swedish) in Stockholm. I will give a talk on our proposed emission limits for antibiotics published last year (the paper is available here), but there will also be talks on wastewater treatment, sustainable pharmaceutical usage and environmental standards for pharmaceuticals. The full program can be found here, and you may register here until June 9. The seminar is free of charge.
And if you are interested in this, I can also recommend the webinar given by Healthcare Without Harm next week (on June 8), which will deal with sustainable procurement as a means to deal with pharmaceutical pollution in the environment. I will at least tune in to hear how the discussion goes here.
Report on JRC AMR workshop
In March, I attended a workshop on the role of NGS technologies in the coordinated action plan against antimicrobial resistance, organised by JRC in Italy. I was, together with 14 other experts, invited to discuss where and how sequencing can be used to investigate and manage antibiotic resistance. The report from the workshop has just recently been published, and is available here. There will be follow-up activities on this workshop, which I also hope that I will be able to participate in, since this is an important and very interesting pet topic of mine.
Reference
Explaining your job to a five-year-old
This morning as I was leaving my daughter at daycare, I got asked by one of the other kids at kindergarten what I do for work. Trying to communicate what you do as a researcher to a five-year-old is a quite interesting task. Five-year-olds are smart – but not very knowledgeable, which leads to very interesting turns to the conversation. Here’s the entire dialogue, transcribed from memory and translated to English:
– What do you do for work?
– I work at the hospital, but I’m not a doctor.
– So you are a psychologist?
– No, I am something called a researcher. I try to understand why bacteria turn evil and make us sick.
– Does someone need to do that?
– Not really. But if we can understand why bugs go bad, we may be able to be sick for much shorter in the future. Or perhaps not get sick at all.
– Okay. Isn’t that hard?
– Yes it is.
– Okay. Bye!
A few things I learned from this conversation: 1) explaining your research to young kids really makes you think about how to present what you do. 2) Kids really question the usefulness of your work (“Does someone need to do that?”). This is actually quite cool, because you need to think about how useful your work really is, in terms that a five-year-old can understand. 3) Society is awesome! To some extent, my work is a “luxury job”, i.e. maybe someone does not need to do my work, but it something we can afford because we share responsibilities and work together as a society, improving (hopefully) the world for all of us. In some sense, nobody strictly needs to be building houses; everyone could just build their own cottage. But building houses improve the standards for everyone, setting time aside for curing diseases, making music, researching microbial interactions, gardening, coffee roasting… Society is awesome.