A thought on peer review and responsibility
I read an interesting note today in Nature regarding the willingness to be review papers. The author of the note (Dan Graur) claims that scientists that publish many papers contribute less to peer review, and proposes a system in which “journals should ask senior authors to provide evidence of their contribution to peer review as a condition for considering their manuscripts.” I think that this is a very interesting thought, however I see other problems coming with it. Let us for example assume that a senior author is neglecting peer review not to be evil, but simply due to an already monumental workload. If we force peer review on such a person, what kind of reviews do we expect to get back? Will this person be able to fulfill a proper, high-quality, peer review assignment? I doubt it.
On the other hand, I don’t have a good alternative either. If no one wants to do the peer reviewing, that system will inevitably break down. However, I think that there would be better to encourage peer review with positive bonuses, rather than pressure – maybe faster handling times, and higher priority, of papers with authors who have done their share of peer reviewing the last two years? Maybe cheaper publishing costs? In any case, I welcome that the subject is brought up for debate, since it is immensely important for the way we perform science today. Thanks Dan!
Regarding ResearchGate and paper requests
I have recently started to receive requests for full-text versions of my publications on ResearchGate. That’s great, but I have yet to figure out how to send them over, without breaking any agreements. As I am in a somewhat intensive work-period at the moment, please forgive me for not spending time on ResearchGate right now. And if you would like full-text versions of my publications, please send me an e-mail! I’ll be glad to help!
ITSx – a software tool for detection and extraction of ITS1 and ITS2 sequences
For a couple of years, I have been working with microbial ecology and diversity, and how such features can be assessed using molecular barcodes, such as the SSU (16S/18S) rRNA sequence (the Metaxa and Megraft packages). However, I have also been aiming at the ITS region, and how that can be used in barcoding (see e.g. the guidelines we published last year). It is therefore a great pleasure to introduce my next gem for community analysis; a software tool for detection and extraction of the ITS1 and ITS2 regions of ITS sequences from environmental communities. The tool is dubbed ITSx, and supersedes the more specific fungal ITS extractor written by Henrik Nilsson and colleagues. Henrik is once more the mastermind behind this completely rewritten version, in which I have done the lion’s share of the programming. Among the new features in ITSx are:
- Robust support for the Cantharellus, Craterellus, and Tulasnella genera of fungi
- Support for nineteen additional eukaryotic groups on top of the already present support for fungi (specifically these groups: Tracheophyta (vascular plants), Bryophyta (bryophytes), Marchantiophyta (liverworts), Chlorophyta (green algae), Rhodophyta (red algae), Phaeophyceae (brown algae), Metazoa (metazoans), Oomycota (oomycetes), Alveolata (alveolates), Amoebozoa (amoebozoans), Euglenozoa, Rhizaria, Bacillariophyta (diatoms), Eustigmatophyceae (eustigmatophytes), Raphidophyceae (raphidophytes), Synurophyceae (synurids), Haptophyceae (haptophytes) , Apusozoa, and Parabasalia (parabasalids))
- Multi-processor support
- Extensive output options
- Virtually zero false-positive extractions
ITSx is today moved from a private pre-release state to a public beta state. No code changes has been made since February, indicative of that the last pre-release candidate is now ready to fly on its own. As far as our testing has revealed, this version seems to be bug free. In reality though, researchers tend to find the most unexpected usage scenarios. So please, if you find any unexpected behavior in this version of ITSx, send me an e-mail and make us aware of the potential shortcomings of our software.
We expect this open-source software to boost research in microbial ecology based on barcoding of the ITS region, and hope that the research community will evaluate its performance also among the eukaryote groups that we have less experience with.
Introducing the PETKit
You know the feeling when your assembler supports paired-end sequences, but your FASTQ quality filterer doesn’t care about what pairs that belong together? Meaning that you end up with a mess of sequences that you have to script together in some way. Gosh, that feeling is way too common. It is for situations like that I have put together the Paired-End ToolKit (PETKit), a collection of FASTQ/FASTA sequence handling programs written in Perl. Currently the toolkit contains three command-line tools that does sequence conversion, quality filtering, and ORF prediction, all adapted for paired-end sequences specifically. You can read more about the programs, which are released as open source software, on the PETKit page. At the moment they lack proper documentation, but running the software with the “–help” option should bring up a useful set of options for each tool. This is still considered beta-software, so any bug reports, and especially suggestions, are welcome.
Also, if you have an idea of another problem that is unsolved or badly executed for paired-end sequences, let me know, and I will see if I can implement it in PETKit.
Published paper: Guidelines for DNA quality checking
I have co-authored a paper together with, among others, Henrik Nilsson that was published today in MycoKeys. The paper deals with checking quality of DNA sequences prior to using them for research purposes. In our opinion, a lot of the software available for sequence quality management is rather complex and resource intensive. Not everyone have the skills to master such software, and in addition computational resources might also be scarce. Luckily, there’s a lot that can be done in quality control of DNA sequences just using manual means and a web browser. This paper puts these means together into one comprehensible and easy-to-digest document. Our targeted audience is primaily biologists who do not have a strong background in computer science, and still have a dataset requiring DNA sequence quality control.
We have chosen to focus on the fungal ITS barcoding region, but the guidelines should be pretty general and applicable to most groups of organisms. In very short our five guidelines spells:
- Establish that the sequences come from the intended gene or marker
Can be done using a multiple alignment of the sequences and verifying that they all feature some suitable, conserved sub-region (the 5.8S gene in the ITS case) - Establish that all sequences are given in the correct (5’ to 3’) orientation
Examine the alignment for any sequences that do not align at all to the others; re-orient these; re-run the alignment step; and examine them again - Establish that there are no (at least bad cases of) chimeras in the dataset
Run the sequences through BLAST in one of the large sequence databases, e.g. at NCBI (or in the ITS case, use the UNITE database), to verify that the best match comprises more or less the full length of the query sequences - Establish that there are no other major technical errors in the sequences
Examine the BLAST results carefully, particularly the graphical overview and the pairwise alignment, for anomalies (there are some nice figures in the paper on how it should and should not look like) - Establish that any taxonomic annotations given to the sequences make sense
Examine the BLAST hit list to see that the species names produced make sense
A much more thorough description of these guidelines can be found in the paper itself, which is available under open access from MycoKeys. There’s simply no reason not to go there and at least take a look at it. Happy quality control!
Reference
Nilsson RH, Tedersoo L, Abarenkov K, Ryberg M, Kristiansson E, Hartmann M, Schoch CL, Nylander JAA, Bergsten J, Porter TM, Jumpponen A, Vaishampayan P, Ovaskainen O, Hallenberg N, Bengtsson-Palme J, Eriksson KM, Larsson K-H, Larsson E, Kõljalg U: Five simple guidelines for establishing basic authenticity and reliability of newly generated fungal ITS sequences. MycoKeys. Issue 4 (2012), 37–63. doi: 10.3897/mycokeys.4.3606 [Paper link]
Improving Swedish research – is there a need for a research elite?
I know that this is not supposed to be a political page, but writing this up, I realized that there is no way I can keep my political views entirely out of this post. So just a quick warning, the following text contains political opinions and is a reflection of my views and believes rather than well supported facts.
So, Swedish minister for education Jan Björklund has announced the government’s plan to spend 3 billion SEK (~350 million EUR, ~450 million USD) on “elite” researchers over the next ten years. One main reason to do so is to strengthen Swedish research in competition with American universities, and to be able to recruit top researchers from other countries to Sweden. While I welcome the prospect of more money to research, I have to say I am very skeptical about the nature of how this money is distributed. First of all, giving more money to the researchers that have already succeeded (I guess this is how you would define elite researchers – if someone has a better idea, please tell both me and Jan Björklund), is not going to generate more innovative research – just more of the same (or similar) things to what these researchers already do. If the government is serious about that Swedish research has a lower-than-expected output (which is a questionable statement in itself), the best way of increasing that output would be to give more researchers the opportunity to put their ideas into action. Second, a huge problem for research in Sweden is that a lot of the scientists’ time is spent on doing other stuff – writing grant applications, administering courses, filling in forms etc. Therefore, one way of improving research would be to put more money into funding at the university administration level, so that researchers actually have time to do what they are supposed to do. I will now provide my own four-point program for how I think that Sweden should move forward to improve the output of science.
1. Researchers need more time
My first point is that researchers need more time to do what they are supposed to do – science. This means that they cannot be expected to apply for money from six different research foundations every year, just to receive a very small amount of money that will keep them from getting thrown out for another 8 months. The short-term contracts that are currently the norm in Sweden create a system where way too much time is spent on writing grant applications – the majority of which will not succeed. In addition, researchers are often expected to be their own secretary, as well as organizing courses (not only lecturing). To solve this we need:
- Longer contracts for scientists. A grant should be large enough to secure five years of salary, plus equipment costs. This allows for some time to actually get the science done, not just the time to write the next application.
- Grants that come with a guaranteed five-year extension of grants to projects that have fulfilled their goals in the first five years. This further secures longevity of researchers and their projects. Also, this allows for universities to actually employ scientists instead of the current system which is all about trying to work around the employment rules.
- More money to university administration. It is simple more cost efficient to have a secretary handling non-science related stuff in the department or group, as well as economic people handling the economy. The current system expects every researcher to be a jack of all trades – which efficiently reduces one to a master of none. More money to administration means more time spent on research.
2. Broad funding creates a foundation for success
Another problem is that if only a few projects are funded repeatedly, the success of Swedish research is very much bound to the success of these projects. While large-scale and high-cost projects are definitely needed, there is also a need to invest in a variety of projects. Many applied ideas have originated from very non-applied research, and the applied research need fundamental research to be done to be able to move forward. However, in the shortsighted governmental view of science, the output has to be almost immediate, which means that applied projects are much more likely to be funded. Thus, projects that could do fundamental discoveries, but are more complicated and take longer time will be down-prioritized by both researchers and universities. To further make situation worse, Björklund et al. have promised more money to universities that cut out non-productive research, with almost guarantees that any projects with a ten-year timeframe will not even be started.
If we are serious about making Swedish research successful, we need to do exactly the opposite. Fund a lot of different projects, both applied and fundamental, regardless of their short-term value. Because the ideas that are most likely to produce short-term results are probably also the ones that are the least innovative in the long-term. Consequently, we need to:
- Spend research funding on a variety of projects, both of fundamental and applied nature.
- Secure funding for “crazy” projects that span long periods of time, at least five to ten years.
3. If we don’t dare to fail, we will not have a chance to win
Finally, research funding must become better at taking risks. If we only bet our money on the most successful researchers, there is absolutely no chance for young scientists to get funded, unless of course they have been picked up by one of the right supervisors. This means that the same ideas get disseminated through the system over and over again, at the expense of more innovative ideas that could pop up in groups with less money to realize them. If these untested ideas in smaller groups get funded, some of them might undoubtedly fail to produce research of high societal value. But some of them will likely develop entirely new ideas, which in the long term might be much more fruitful than throwing money on the same groups over and over again. Suggestions:
- Spend research funding broadly and with an active risk-gain management strategy.
- Allow for fundamental research to investigate completely new concepts – even if they are previously untested, and regardless (or less dependent on) previous research output.
- Invest in infrastructure for innovative research – and do so fast. For example, the money spent on the sequencing facilities at Sci Life Lab in Stockholm is an excellent example of an infrastructure investment that gains a lot of researchers at different universities access to high-throughput sequencing, without each university having to invest in expensive sequencing platforms themselves. More such centers would both spur collaboration and allow for faster adoption of new technologies.
4. Competing with what we are best at
A mistake that is often done when trying to compete with those that are best in the class is to try to compete by doing the same things as the best players do. This makes it extremely hard to win a game against exactly those players, as they are likely more experienced, have more resources, and already has the attention to get the resources we compete for. Instead, one could try to play the Wayne Gretzky trick: to try to skate where the puck is heading, instead of where it is today. Another approach would be to invent a new arena for the puck to land in, where you have better control over the settings than your competitors (slightly similar to what Apple did when the iPod was released, and Microsoft couldn’t use Windows to leverage their mp3-player Zune).
For Sweden, this would mean that we should not throw some bucks at the best players at our universities and hope that they will be happy with this (comparably small) amount of money. Instead, we should give them circumstances to work under that are much better or appealing from other standpoints. This could be better job security, longer contracts, less administrative work, securer grants, more freedom to decide over ones time, and larger possibilities to combine work and family. Simply creating a better, securer and nicer environment to work in. However, Björklund’s suggestions go the very opposite way: researchers should compete to be part of the elite community, and if your not in that group, you’d get thrown out. Therefore, I suggest (with the risk of repeating myself) that we should compete by:
- Offering longer contracts and grants for scientists.
- Giving scientists opportunities to combine work and family life.
- Embracing all kinds of science, both fundamental and applied, both short-term and long-term.
- Allowing researchers to take risks, even if they fail.
- Giving universities enough funding to let scientists do the science and administrative personal do the administration.
- Funding large-scale collaborative infrastructure investments.
- Thinking of how to create an environment that is appealing for scientists, not only from an economic perspective.
A note on other important aspects of funding
Finally, I have now been focusing a lot on width as opposed to directed funding to an elite research squad. It is, however, apparent that we also need to allocate funding to bring in more women to the top positions in the academy. Likely, a system which favors elite groups will also favor male researchers, judging from how the Swedish Foundation for Strategic Research picks their bets for the future. Also, it is important that young researchers without strong track records gets funded, otherwise a lot of new and interesting ideas risk to be lost.
In the fourth point of my proposal, I suggest that Sweden should compete at what Sweden is good at, that is to view researchers as human beings, which are most likely to succeed in an environment where they can develop their ideas in a free and secure way. For me, it is surprising that a minister of education representing a liberal party wants to excess such control over what is good and bad research. Putting up a working social security system around science seems much more logical than throwing money at those who already have. Apparently I have forgotten that our current government is not interested in having a working social security system – their interest seem to lie in deconstructing the very same structures.
Pfam team aims at cleaning erroneous protein families
The guys at Pfam recently introduced a new database, called AntiFam, which will provide HMM profiles for some groups of sequences that seemingly formed larger protein families, although they were not actually real proteins. For example, rRNA sequences could contain putative ORFs, that seems to be conserved over broad lineages; with the only problem being that they are not translated into proteins in real life, as they are part of an rRNA [1].
With this initiative the Xfam team wants to “reduce the number of spurious proteins that make their way into the protein sequence databases.” I have run into this problem myself at some occasions with suspicious sequences in GenBank, and I highly encourage this development towards consistency and correctness in sequence databases. It is of extreme importance that databases remain reliable if we want bioinformatics to tell us anything about organismal or community functions. The Antifam database is a first step towards such a cleanup of the databases, and as such I would like to applaud Pfam for taking actions in this direction.
To my knowledge, GenBank are doing what they can with e.g. barcoding data (SSU, LSU, ITS sequences), but for bioinformatics and metagenomics (and even genomics) to remain viable, these initiatives needs to come quickly; and automated (but still very sensitive) tools for this needs to get our focus immediately. For example, Metaxa [2] could be used as a tool to clean up SSU sequences of misclassified origin. More such tools are needed, and a lot of work remains to be done in the area of keeping databases trustworthy in the age of large-scale sequencing.
References
- Tripp, H. J., Hewson, I., Boyarsky, S., Stuart, J. M., & Zehr, J. P. (2011). Misannotations of rRNA can now generate 90% false positive protein matches in metatranscriptomic studies. Nucleic Acids Research, 39(20), 8792–8802. doi:10.1093/nar/gkr576
- Bengtsson, J., Eriksson, K. M., Hartmann, M., Wang, Z., Shenoy, B. D., Grelet, G.-A., Abarenkov, K., et al. (2011). Metaxa: a software tool for automated detection and discrimination among ribosomal small subunit (12S/16S/18S) sequences of archaea, bacteria, eukaryotes, mitochondria, and chloroplasts in metagenomes and environmental sequencing datasets. Antonie van Leeuwenhoek, 100(3), 471–475. doi:10.1007/s10482-011-9598-6
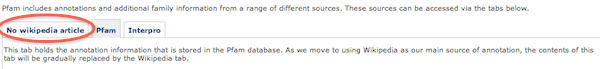
Pfam + Wikipedia – finally!
Browsing the Pfam web site today, I discovered that the database finally has launched its Wikipedia co-ordination efforts.
This has happened along with the 25th release of the Pfam database (released 1st of April), and basically means that Wikipedia articles will be linked to Pfam families. Gradually, this will (hopefully) improve the annotation of Pfam families, which has in many cases been rather poor. The Xfam blog post related to Pfam release 25 says the change will be happening gradually, which might actually be good thing, given the quirks that might pop up.
(…) a major change is that Pfam annotation is now beginning to be co-ordinated via Wikipedia. Unlike Rfam, where every entry has a Wikipedia entry, we expect this to be a more gradual transition for Pfam, so not all entries currently have a corresponding Wikipedia article. For a more detailed discussion, check the help page. We actively encourage the addition of new/updated annotations via Wikipedia as they will appear far quicker than waiting for a Pfam release. If there are articles in Wikipedia that you think correspond to a family, then please mail us!
I have awaited this change for a long time, and is very happy that Pfam has finally taken this step. Congratulations and my sincerest thanks to the Pfam team! Now, let’s go editing!
Underpinning Wikipedia’s Wisdom
In December, Alex Bateman, whose opinions on open science I support and have touched upon earlier, wrote a short correspondence letter to Nature [1] in which he again repeated the points of his talk at FEBS last summer. He concludes by the paragraph:
Many in the scientific community will admit to using Wikipedia occasionally, yet few have contributed content. For society’s sake, scientists must overcome their reluctance to embrace this resource.
I agree with this statement. However, as I also touched upon earlier, but like to repeat again – bold statements doesn’t make dreams come true – action does. Rfam, and the collaboration with RNA Biology and Wikipedia is a great example of such actions. So what other actions may be necessary to get researchers to contribute to the Wikipedian wisdom?
First of all, I do not think that the main obstacle to get researchers to edit Wikipedia articles is reluctance to doing so because Wikipedia is “inconsistent with traditional academic scholarship”, though that might be a partial explanation. What I think is the major problem is the time-reward tradeoff. Given the focus on publishing peer-reviewed articles, the race for higher impact factor, and the general tendency of measuring science by statistical measures, it should be no surprise that Wikipedia editing is far down on most scientists to-do lists, so also on mine. The reward of editing a Wikipedia article is a good feeling in your stomach that you have benefitted society. Good stomach feelings will, however, feed my children just as little as freedom of speech. Still, both Wikipedia editing and freedom of speech are extremely important, especially as a scientist.
Thus, there is a great need of a system that:
- Provides a reward or acknowledgement for Wikipedia editing.
- Makes Wikipedia editing economically sustainable.
- Encourages publishing of Wikipedia articles, or contributions to existing ones as part of the scientific publishing process.
Such a system could include a “contribution factor” similar to the impact factor, in which contribution of Wikipedia and other open access forums was weighted, with or without a usefulness measure. Such a usefulness measure could easily be determined by links from other Wikipedia articles, or similar. I realise that there would be severe drawbacks of such a system, similar to those of the impact factor system. I am not a huge fan of impact factors (read e.g. Per Seglen’s 1997 BMJ article [2] for some reasons why), but I do not see that system changing any time soon, and thus some kind of contribution factor could provide an additional statistical measure for evaluators to consider when examining scientists’ work.
While a contribution factor would be an incitement for researchers to contribute to the common knowledge, it will still not provide an economic value to do so. This could easily be changed by allowing, and maybe even requiring, scientists to contribute to Wikipedia and other public fora of scientific information as part of their science outreach duties. In fact, this public outreach duty (“tredje uppgiften” in Swedish) is governed in Swedish law. In 2009, the universities in Sweden have been assigned to “collaborate with the society and inform about their operations, and act such that scientific results produced at the university benefits society” (my translation). It seems rational that Wikipedia editing would be part of that duty, as that is the place were many (most?) people find information online today. Consequently, it is only up to the universities to demand 30 minutes of Wikipedia editing per week/month from their employees. Note here that I am referring to paid editing.
Another way of increasing the economic appeal of writing Wikipedia articles would be to encourage funding agencies and foundations to demand Wikipedia articles or similar as part of project reports. This would require researchers to make their findings public in order to get further funding, a move that would greatly increase the importance of increasing the common wisdom treasure. However, I suspect that many funding agencies, as well as researchers would be reluctant to such a solution.
Lastly, as shown by the Rfam/RNA Biology/Wikipedia relationship, scientific publishing itself could be tied to Wikipedia editing. This process could be started by e.g. open access journals such as PLoS ONE, either by demanding short Wikipedia notes to get an article published, or by simply provide prioritised publishing of articles which also have an accompanying Wiki-article. As mentioned previously, these short Wikipedia notes would also go through a peer-review process along with the full article. By tying this to the contribution factor, further incitements could be provided to get scientific progress in the hands of the general public.
Now, all these ideas put a huge burden on already hard-working scientists. I realise that they cannot all be introduced simultaneously. Opening up publishing requires time and thought, and should be done in small steps. But doing so is in the interest of scientists, the general public and the funders, as well as politicians. Because in the long run it will be hard to argue that society should pay for science when scientists are reluctant to even provide the public with an understandable version of the results. Instead of digging such a hole for ourselves, we should adapt the reward, evaluation, funding and publishing systems in a way that they benefit both researchers and the society we often say we serve.
- Bateman and Logan. Time to underpin Wikipedia wisdom. Nature (2010) vol. 468 (7325) pp. 765
- Seglen. Why the impact factor of journals should not be used for evaluating research. BMJ (1997) vol. 314 (7079) pp. 498-502
Alex Bateman says protein family wiki is coming – but when?
Nature recently had a nice news article on Bio-wikis and biological databases connected to Wikipedia where Alex Bateman says they’re working on a protein-family wiki that will be hosted on Wikipedia, similar to the Rfam wiki, which he talked about at FEBS this summer. I am of course very excited about this, and hope that the new Pfam (?) wiki will come rather sooner than later. As pointed out earlier, the Nature article also underlines the problem of scientific wikis; currently there is no career incentive to get researchers to spend their time editing wiki-articles. That is a shame, and perhaps a system copying the system of Rfam and RNA Biology could help in this direction. The only question is which journal(s) that would be interested in such a commitment to open science…