My ISME talk on EMBARK
Ákos Kovács had the brilliant idea of putting up a temporary resource for things you bring up in a talk that you can point people to. I did not do this before my talk at ISME today, but I thought the idea was so good, so here’s a summary and collection of my ISME short-talk on the EMBARK outcomes today:
- More information on EMBARK and its successor SEARCHER can be found on the project website, here: http://antimicrobialresistance.eu Importantly, this is a team effort over four years and I only touched on a few selected things
- Within the project we have looked at typical background levels of antibiotic resistance in the environment. We have already published some of these results (for qPCR abundances) in Abramova et al. 2023
- The average resistance gene in the average environment is present in ~1 in 1000 bacteria, but the variation between different genes is huge
- Depending on monitoring goal, different target genes are relevant to use. See this table adapted from Abramova et al. 2023:
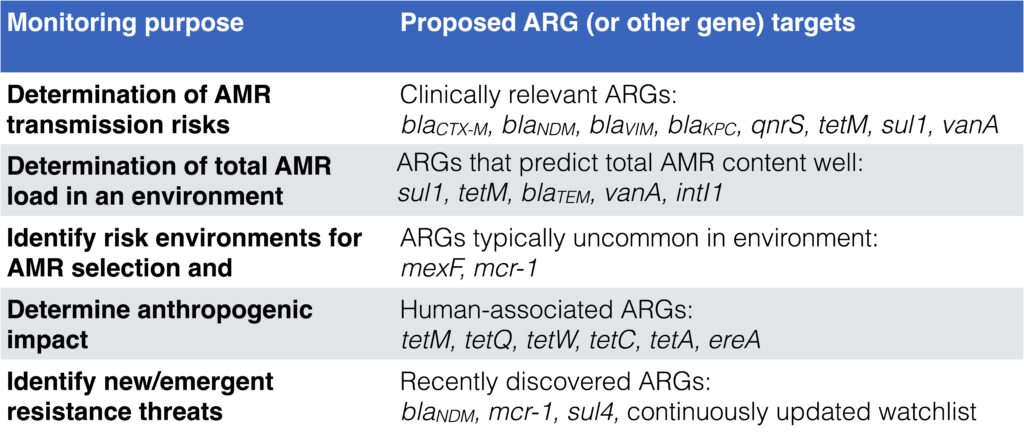
- We have also tried to make different monitoring methods for environmental AMR comparable. Those mentioned in the talk were selective culturing for resistant bacteria, qPCR and shotgun metagenomics
- This data is not yet published, but overall we see relatively good correlation between qPCR and metagenomics. This is not true for all genes, though, and unfortunately neither qPCR nor metagenomics is always better than the other
- Culturing data is not very good at predicting specific antibiotic resistance gene abundances as the class level
- Finally, we have developed methods for discovering new types of ARGs, as seen in the ResFinderFG database: Gschwind et al. 2023
- We have also used these new methods to look at differences between established ARGs and latent ARGs in a variety of environments: Inda-Díaz et al. 2023
- Our ultimate goal in EMBARK would be to develop a modular framework for environmental monitoring of antibiotic resistance. You can read more about our thinking and goals in the review paper we published last year: Bengtsson-Palme et al. 2023
Published paper: E. coli in coastal marine sediments
Last week, FEMS Microbes published our most recent work on the genomes of Escherichia coli in coastal marine sediments from the Helsingborg area in Sweden (1). Part of our sampled area was next to the discharge point of the city’s wastewater treatment plant (WWTP) effluent. We discovered that the E. coli population in these sediment is diverse, containing serotypes typically associated with both humans, livestock and other animals. We also found that virulence genes were more common among the isolates collected closer to the WWTP discharge site. Only one isolate was phenotypically antibiotic resistant, and carried corresponding tetracycline resistance genes on a plasmid. All isolates were halotolerant, growing at 3.5% NaCl. Since most isolates were also good at forming biofilm, this suggests that marine sediments can select for E. coli with increased survival properties and could be a potential reservoir for E. coli that could be spread to humans when the sediments are disturbed. Furthermore, the naturalisation of these E. coli questions it as an indicator for faecal contamination of marine sediments.
The paper is primarily the work of Isabel Erb, Carolina Suarez, and Catherine Paul at Lund University, and they have made a terrific job on this while I have mostly provided some input on the bioinformatics and genomics analyses. The study is a nice example of how genomics analysis could nuance monitoring for pathogens and antibiotic resistance in environments close to human activities. Since these sediments are also closely connected to humans in terms of exposure – the Helsingborg beach is in the neighbouring area – this highlight potential exposure routes for pathogens and antibiotic resistance (2).
The finding of a single antibiotic resistant isolate highlights the issue of comparing between different monitoring methods (2). While a single isolates might be consider a small number, it is really hard to compare if this is outside of the normal range of resistance (3) as measured by, e.g., qPCR. This further points to the importance of standardisation of antibiotic resistance monitoring in the environment, in a way that is both reliable, feasible and economic. That said, it also shows the potential in monitoring, for example, public beaches for pathogens and resistance, and how this could be used to better design and implement mitigation strategies, including the temporary closing of public beaches in contaminated areas. For this to work, however, a better knowledge of the background levels of resistance is required, as we have been working on in the EMBARK program.
References
- Erb IK, Suarez C, Frank EM, Bengtsson-Palme J, Lindberg E, Paul CJ: Escherichia coli in urban marine sediments: interpreting virulence, biofilm formation, halotolerance and antibiotic resistance to infer contamination or naturalisation. FEMS Microbes (advance article) xtae024 (2024). doi: 10.1093/femsmc/xtae024
- Bengtsson-Palme J, Abramova A, Berendonk TU, Coelho LP, Forslund SK, Gschwind R, Heikinheimo A, Jarquin-Diaz VH, Khan AA, Klümper U, Löber U, Nekoro M, Osińska AD, Ugarcina Perovic S, Pitkänen T, Rødland EK, Ruppé E, Wasteson Y, Wester AL, Zahra R: Towards monitoring of antimicrobial resistance in the environment: For what reasons, how to implement it, and what are the data needs? Environment International, 178, 108089 (2023). doi: 10.1016/j.envint.2023.108089
- Abramova A, Berendonk TU, Bengtsson-Palme J: A global baseline for qPCR-determined antimicrobial resistance gene prevalence across environments. Environment International, 178, 108084 (2023). doi: 10.1016/j.envint.2023.108084
Published paper: Aberrant microbiomes in mice and increased antibiotic resistance
This paper came out just about the same time as the PhD position with Erik Kristiansson where I will be co-supervisor was announced, and I did not want to steal that thunder with another news item, but it is now time to highlight the fantastic work of Víctor Hugo Jarquín-Díaz on antibiotic resistance genes in the gut microbiomes of mice across a gradient of pure and hybrid genotypes in the European house mouse hybrid zone. This came out in mid-April in ISME Communications and presents the interesting hypothesis that hybridisation not only shapes bacterial communities, but also antibiotic resistance gene occurrences (1).
This study is based on 16S rRNA amplicon sequencing of gut bacteria in natural populations of house mice. From this we have predicted the antibiotic resistance gene composition in the microbiomes, and found a significant increase in the predicted antibiotic resistance gene richness in hybrid mice. In other words, more and different antibiotic resistance genes were found in the hybrid mice than in the non-hybridised individuals. We believe that this could be due to a disruption of the microbiome composition in hybrid mice. The aberrant microbiomes in hybrids represent less complex communities, potentially promoting selection for resistance.
It deserves to be mentioned that this is more of a pilot study, which we hope to follow up with a more proper study targeting the resistance genes in the mice microbiomes. That said, our work suggests that host genetic variation impacts the gut microbiome and antibiotic resistance gene, at least in mice. This raises further questions on how the mammalian host genetics impact antibiotic resistance carriage in bacteria via microbiome dynamics or interaction with the environment.
I am very happy to have been part of this EMBARK collaboration with the Sofia Forslund-Startceva and Emanuel Heitlinger labs! And I am especially thankful to Víctor who pulled off this very thought-inducing study!
Reference
- Jarquín-Díaz VH, Ferreira SCM, Balard A, Ďureje Ľ, Macholán M, Piálek J, Bengtsson-Palme J, Kramer-Schadt S, Forslund-Startceva SK, Heitlinger E: Aberrant microbiomes are associated with increased antibiotic resistance gene load in hybrid mice. ISME Communications, ycae053 (2024). doi: 10.1093/ismeco/ycae053 [Paper link]
Welcome back Agata
I am very happy to welcome Agata Marchi back to the group as a PhD student! Agata was a master student in the group last year, doing a thesis focused on implementing a bioinformatic approach to identify differences between the genomes of host-associated and non host-associated strains of Pseudomonas aeruginosa. While one of her first tasks will be to complete this work and prepare it for publication, her doctoral studies will primarily be on the interactions between bacteria and between bacteria and host in the human microbiome and how these relate to complex diseases. She will focus on developing and applying machine learning methods to better understand this interplay.
I am – as the rest of the group – very happy to welcome Agata back to the lab!
Conferences this fall
Time to do a rundown of conferences and meetings I will attend this fall. Double-check with your calendars and please reach out if you’re also going, so we can meet up!
September 21-24: Nordic Society of Clinical Microbiology and Infectious Diseases (NSCMID), in Örebro, Sweden. I will give a talk about the EMBARK work in the Saturday session on Metagenomics in infection, inflammatory disease and the environment
October 5-6: Conference on ‘Optimal practices to protect human health care from antimicrobial resistance selected in the veterinary domain’ organised by The Netherlands Food and Consumer Product Safety Authority (NVWA) in Amsterdam, the Netherlands. I will chair a session on October 6 on Next generation sequencing for bioinformatic based surveillance.
October 18-22: 32º Congresso Brasileiro de Microbiologia, in Foz do Iguaçu, Brazil. I will give a talk in the Saturday session (the 21st) on the use of model systems all the way to global surveillance systems to prevent future pandemics.
November 15-16: DDLS Annual Meeting, in Stockholm Sweden. I am in the organising committee for this event with the theme “The emerging role of AI in data-driven life science”.
November 17: DDLS Cell and Molecular Biology Minisymposium.
November 29: GOTBIN Annual Workshop, in Gothenburg Sweden.
This will be a fun (but intense!) fall!
PhD position with Luis Pedro Coelho
I just want to point potential doctoral students’ attention to this fantastic opportunity to work with my EMBARK colleague Luis Pedro Coelho as he sets up his new lab in Brisbane in Australia at the relatively new Centre for Microbiome Research. Luis is looking for two PhD students, one who will focus on identifying and characterising the small proteins of the global microbiome and one more related to developing novel bioinformatic methods for studying microbial communities.
I can highly recommend this opportunity given that you are willing to move to Australia, as Luis is one of the most brilliant scientists I have worked with, is incredibly easy-going and fosters a lab culture I strong support. More information and application here.
ITSx in Galaxy
I am happy to share with you that since a couple of months back there is an up-to-date version of ITSx available through Galaxy! The tool can be found here: https://usegalaxy.eu/root?tool_id=itsx
The person behind this is really Björn Grüning at the University of Freiburg. I am immensely thankful for the work he has put into this. Our intention to make sure that both the Galaxy version and the bioconda version are maintained in parallel to the one on this website, and continuously up to date!
Happy barcoding!
New team members
Time is passing quickly, and I have not appropriately acknowledged the many newcomers we’ve had to the lab in the past couple of months. With this post I would like to say welcome to the lab to Máté Vass and Dani Jáen Luchoro (both postdocs), Jorge Agramont and Josue Mamani Jarro (doctoral students), as well as Nathália Abichabki (visiting doctoral student from Brazil)! Some of you have already spent a couple of months in the group and we very much enjoy having you here!

A week or so ago, we took this new lab picture with everyone (except for Lisa, who is in Amsterdam). I am very proud to be working with group of extremely talented, smart, funny and goodhearted people!
Very briefly, Dani will be working on updating the BacMet database as part of the BIOCIDE project, and shares his time between my group, Joakim Larsson‘s group and the Sahlgrenska hospital. Máté was recruited within the DDLS program and will work on inferring the metacommunity ecology of antibiotic resistance based on analysis of large-scale datasets. Jorge and Josue are part of the same SIDA-funded doctoral student exchange program with Bolivia and will work on different aspects of environmental antibiotic resistance and the spread of diarrheal pathogens through the environmental matrix. Nathália, finally, is working on understanding the tolerance mechanisms to antibiotics in Klebsiella pneumoniae.
All of you are very welcome to the group!
Published paper: Preterm infant microbiome and resistome
Together with our collaborators in Tromsø in Norway, we published a paper over the weekend in eBioMedicine describing the early colonization patterns of preterm infants, both in terms of the microbes that arrive early to the infants, but also in terms of the antibiotic resistance genes they carry.
In the paper (1), which is a continuation of an earlier study by part of the team (2), we analysed metagenomic data from six Norwegian neonatal intensive care units to better understand the bacterial microbiota of infants born preterm or on term and receiving different treatments. These groups included probiotic-supplemented and antibiotic-exposed extremely preterm infants (n = 29), antibiotic-exposed very preterm infants (n = 25), antibiotic-unexposed very preterm infants (n = 8), and antibiotic-unexposed full-term infants (n = 10). Stool samples were collected from the infants after 7, 28, 120, and 365 days of life and were analysed using shotgun metagenomics. We were particularly interested in the maturation of the preterm infant microbiome into a ‘normal’ healthy gut microbiome, and the colonization with bacteria carrying antibiotic resistance genes.
We found that microbiota maturation was largely determined by the length of hospitalisation for the infants and how much preterm they were. The use of probiotics rendered the gut microbiota and resistome of extremely preterm infants more alike to term infants on day 7 and partially restored the loss of species interconnectivity and stability associated with preterm delivery. Finally, colonisation with Escherichia coli was associated with the highest number of antibiotic-resistance genes in the infant microbiomes, followed by Klebsiella pneumoniae and Klebsiella aerogenes.
Being born very preterm, along with prolonged hospitalisation and frequent antibiotic use alters early life resistome and mobilome, leading to an increased gut carriage of antibiotic resistance genes and mobile genetic elements. On the other hand, the effect of probiotics was not unidirectional. Probiotics decreased resistome burden, but at the same time the bacterial strains in the probiotics appear to promote the activity of mobile genetic elements. Here, further study of the gut microbiota is necessary to be able to design strategies aiming to lower disease risk in vulnerable preterm infants.
As mentioned, this study was a collaboration with Veronika Pettersen‘s group in Tromsø, particularly Ahmed Bargheet, who have done a fabulous job on the bioinformatics and analysis of this study. I hope that we will continue this collaboration in the future (first step will be me visting Tromsø again in June!) This also continues a nice little “sidetrack” of the group’s research into the early life microbiome – previously represented by the work of Katariina Pärnänen (3) and Tove Wikström‘s vaginal microbiome study (4), which is a very interesting and relevant subject in terms of both medicine and microbial ecology. We are also setting up new collaborations in this area, so I hope that more will come out of this track in the next couple of years.
Finally, thank you Veronika for inviting me to participate in this great project!
References
- Bargheet A, Klingenberg C, Esaiassen E, Hjerde E, Cavanagh JP, Bengtsson-Palme J, Pettersen VK: Development of early life gut resistome and mobilome across gestational ages and microbiota-modifying treatments. eBio Medicine, 92, 104613 (2023). doi: 10.1016/j.ebiom.2023.104613
- Esaiassen E, Hjerde E, Cavanagh JP, Pedersen T, Andresen JH, Rettedal SI, Støen R, Nakstad B, Willassen NP, Klingenberg C: Effects of Probiotic Supplementation on the Gut Microbiota and Antibiotic Resistome Development in Preterm Infants. Frontiers in Pediatrics, 16, 6, 347 (2018). doi: 10.3389/fped.2018.00347
- Pärnänen K, Karkman A, Hultman J, Lyra C, Bengtsson-Palme J, Larsson DGJ, Rautava S, Isolauri E, Salminen S, Kumar H, Satokari R, Virta M: Maternal gut and breast milk microbiota affect infant gut antibiotic resistome and mobile genetic elements. Nature Communications, 9, 3891 (2018). doi: 10.1038/s41467-018-06393-w
- Wikström T, Abrahamsson S, Bengtsson-Palme J, Ek CJ, Kuusela P, Rekabdar E, Lindgren P, Wennerholm UB, Jacobsson B, Valentin L, Hagberg H: Microbial and human transcriptome in vaginal fluid at midgestation: association with spontaneous preterm delivery. Clinical and Translational Medicine, 12, 9, e1023 (2022). doi: 10.1002/ctm2.1023
Emil’s halftime
Some good news from the lab! Emil Burman today passed his halftime control, which means that we now can look forward to around more years of fun science together! We all congratulate Emil on this great achievement which marks an important milestone in the group, as Emil is the first of the PhD students who have reached it to this point!