Summary of 2016
 What a year it has been. Let’s just say that this year brought a lot of interesting stuff – some good, some bad. On a personal level, mostly good though. First and foremost, I successfully defended my PhD thesis in May. And second, I secured funding for a postdoctoral stay in the US, beginning next summer. In addition, I have been involved in eight published research papers, and three more perspective oriented opinion pieces. But let’s take it from the start.
What a year it has been. Let’s just say that this year brought a lot of interesting stuff – some good, some bad. On a personal level, mostly good though. First and foremost, I successfully defended my PhD thesis in May. And second, I secured funding for a postdoctoral stay in the US, beginning next summer. In addition, I have been involved in eight published research papers, and three more perspective oriented opinion pieces. But let’s take it from the start.
A lot of the research I have published this year have been related to the release of pharmaceuticals, particularly antibiotics, into the environment, and the minimal selective concentrations (MSCs) that can drive antibiotic resistance development. Late 2015 me and Prof. Joakim Larsson described predicted no-effect concentrations (PNECs, in most respect similar to MSCs) for 111 antibiotics, and the Larsson lab built on this publication this year by validating (pretty much at least) the MSC for tetracycline to be around 1 µg/L in complex bacterial communities. We also used these PNECs as a backdrop to better understand the selective potential of antibiotics in Swedish sewage treatment plants, but found that the selective effects of antibiotics on gross resistance gene content seem to be almost negligible compared to the effects caused by other abiotic factors, particularly oxygen availability. That does, of course, not exclude that there might be minor selective effects, but if such effects exist, the metagenomics approaches we used was not sensitive enough to detect them. Me and Joakim also wrote two opinion pieces on selective effects and antibiotics releases from pharmaceutical production, arguing for that the industry should take their responsibility to clean up their act. Still, legislative action towards sustainable production is also needed to as far as possible avoid and delay resistance development in human pathogens.
In the realm of taxonomy, I have contributed to an annotation effort of fungi from the built environment – based on a workshop held in May in Gothenburg. I have also participated in Rodney Richardson’s evaluation of the performance of different software tools for taxonomic classification when used with barcoding sequences outside of the rRNA genes and the ITS region, specifically those used for barcoding of plants. We here found that there is a substantial tradeoff that leads to that tools with high sensitivity also have a high error rate. We think that this could be alleviated with smarter algorithms, which I hope that we will return to in the (relatively near) future.
We also published a paper describing the Metaxa2 Diversity Tools released last year, which ease diversity analyses with Metaxa2. Also, the paper describing FARAO – the Flexible All-Round Annotation Organizer – was published in August in Bioinformatics. Additionally, a paper we started writing three years ago on strategies to improve sequence databases and particularly their annotation finally saw the light of day this year in the journal Proteomics.
Finally, one of the most exciting projects I’ve been part of published this year was the large metagenome screening for resistance genes first-authored by Chandan Pal (who is defending his thesis now in January), in which we identify that environments subjected to pharmaceutical pollution and waste-water treatment plants are the only environments with higher abundances of antibiotic resistance genes than the human and animal guts. However, the largest diversity of resistance genes was found in air from a Beijing smog event, opening up the possibility that this could be an under-investigated dissemination route for resistance genes and resistant bacteria. This finding also fuelled an overly scared news reaction in China (and to some degree understandably so). However, the human health risks associated with this finding is likely to be minute, at least in the short term, compared to all other health problems related to smog.
 In May, I successfully defended my doctoral thesis, which was largely a pleasant experience, at least compared to actually writing it. Particularly complicating things was the fact that the thesis hand-in deadline and the FORMAS post-doc grant application deadline happened to end up on the same time. This dual writing process was quite stressful to say the least. On the other hand, this effort paid of – in November I got a positive response from FORMAS, so I am now heading to Wisconsin later this year to work with Prof. Jo Handelsman, who will start setting up her new lab there in February.
In May, I successfully defended my doctoral thesis, which was largely a pleasant experience, at least compared to actually writing it. Particularly complicating things was the fact that the thesis hand-in deadline and the FORMAS post-doc grant application deadline happened to end up on the same time. This dual writing process was quite stressful to say the least. On the other hand, this effort paid of – in November I got a positive response from FORMAS, so I am now heading to Wisconsin later this year to work with Prof. Jo Handelsman, who will start setting up her new lab there in February.
In relation to the grant writing I paid a short visit to Yale, where Prof. Handelsman was working at the time (when she was not working for President Obama at the White House, that is). It was real fun to meet this incredible (but currently quite small) research group and see the impressive environments at Yale. I participated in the NDPIA meeting in Gothenburg and a conference on metal and biocide co-selection for antibiotic resistance in Stockholm during the spring, and in November I went to London to the Next Generation Sequencing Congress (which this year was a disappointment). Sadly, I missed out of the Swedish Bioinformatics Workshop due to my doctoral promotion ceremony, but I guess sometimes you have to prioritise.
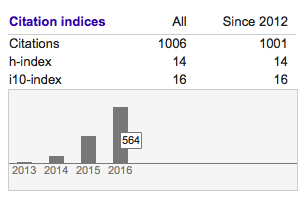 Since I am also obsessed with numbers, I am very happy to report that in late December 2016 I passed 1000 citations according to Google Scholar (for whatever that’s worth…) I am most amazed, however, that I got 564 citations in 2016 alone, meaning that more than one researcher per day actually found some of my work citation-worthy. I am honoured, thank you for using our work in so many different ways!
Since I am also obsessed with numbers, I am very happy to report that in late December 2016 I passed 1000 citations according to Google Scholar (for whatever that’s worth…) I am most amazed, however, that I got 564 citations in 2016 alone, meaning that more than one researcher per day actually found some of my work citation-worthy. I am honoured, thank you for using our work in so many different ways!
In the coming year, I will first of all need to clean up my act with the Joakim Larsson group before leaving for the US. This means writing manuscripts and support other people’s manuscript-writing. We have some co-resistance related work in which I am involved in the late stages of the pipeline, and also some stuff on predicting novel resistance genes. In addition, we are still doing work on minimal selective concentrations, some of which I have taken part in. I have also almost finished a number of review manuscripts which are to be submitted during the next month or so. And of course there is always some work to do on Metaxa2 and ITSx. Except a major ITSx update during the winter or spring, and a major Metaxa2 update at some point later during the spring (although the latter estimate here may be optimistic). I think 2017 will be a very exciting year, and I hope that you will find it exciting too!
Publications
Papers
- Richardson RT, Bengtsson-Palme J, Johnson RM: Evaluating and Optimizing the Performance of Software Commonly Used for the Taxonomic Classification of DNA Sequence Data. Molecular Ecology Resources, Early view (2016). doi: 10.1111/1755-0998.12628 [Paper link]
- Pal C, Bengtsson-Palme J, Kristiansson E, Larsson DGJ: The structure and diversity of human, animal and environmental resistomes. Microbiome, 4, 54 (2016). doi: 10.1186/s40168-016-0199-5 [Paper link]
- Abarenkov K, Adams RI, Laszlo I, Agan A, Ambrioso E, Antonelli A, Bahram M, Bengtsson-Palme J, Bok G, Cangren P, Coimbra V, Coleine C, Gustafsson C, He J, Hofmann T, Kristiansson E, Larsson E, Larsson T, Liu Y, Martinsson S, Meyer W, Panova M, Pombubpa N, Ritter C, Ryberg M, Svantesson S, Scharn R, Svensson O, Töpel M, Untersehrer M, Visagie C, Wurzbacher C, Taylor AFS, Kõljalg U, Schriml L, Nilsson RH: Annotating public fungal ITS sequences from the built environment according to the MIxS-Built Environment standard – a report from a May 23-24, 2016 workshop (Gothenburg, Sweden). MycoKeys, 16, 1–15 (2016). doi: 10.3897/mycokeys.16.10000 [Paper link]
- Bengtsson-Palme J, Hammarén R, Pal C, Östman M, Björlenius B, Flach C-F, Kristiansson E, Fick J, Tysklind M, Larsson DGJ: Elucidating selection processes for antibiotic resistance in sewage treatment plants using metagenomics. Science of the Total Environment, 572, 697–712 (2016). doi: 10.1016/j.scitotenv.2016.06.228 [Paper link]
- Hammarén R, Pal C, Bengtsson-Palme J: FARAO: The Flexible All-Round Annotation Organizer. Bioinformatics, advance access (2016). doi: 10.1093/bioinformatics/btw499 [Paper link]
- Bengtsson-Palme J, Thorell K, Wurzbacher C, Sjöling Å, Nilsson RH: Metaxa2 Diversity Tools: Easing microbial community analysis with Metaxa2. Ecological Informatics, 33, 45–50 (2016). doi: 10.1016/j.ecoinf.2016.04.004 [Paper link]
- Lundström S, Östman M, Bengtsson-Palme J, Rutgersson C, Thoudal M, Sircar T, Blanck H, Eriksson KM, Tysklind M, Flach C-F, Larsson DGJ: Minimal selective concentrations of tetracycline in complex aquatic bacterial biofilms. Science of the Total Environment, 553, 587–595 (2016). doi: 10.1016/j.scitotenv.2016.02.103 [Paper link]
- Bengtsson-Palme J, Larsson DGJ: Why limit antibiotic pollution? The role of environmental selection in antibiotic resistance development. APUA Newsletter, 34, 2, 6-9 (2016). [Paper link].
- Bengtsson-Palme J, Boulund F, Edström R, Feizi A, Johnning A, Jonsson VA, Karlsson FH, Pal C, Pereira MB, Rehammar A, Sánchez J, Sanli K, Thorell K: Strategies to improve usability and preserve accuracy in biological sequence databases. Proteomics, 16, 18, 2454–2460 (2016). doi: 10.1002/pmic.201600034 [Paper link]
- Bengtsson-Palme J, Larsson DGJ: Time to limit antibiotic pollution. The Medicine Maker, 0416, 302, 17–18 (2016). [Paper link]
- Bengtsson-Palme J, Larsson DGJ: Concentrations of antibiotics predicted to select for resistant bacteria: Proposed limits for environmental regulation. Environment International, 86, 140–149 (2016). doi: 10.1016/j.envint.2015.10.015 [Paper link] (Published in late 2015, but noted as a 2016 paper.)
Doctoral thesis
- Bengtsson-Palme J: Antibiotic resistance in the environment: a contribution from metagenomic studies. Doctoral thesis (medicine), Department of Infectious Diseases, Institute of Biomedicine, Sahlgrenska Academy, University of Gothenburg, 2016.
Posters
- Bengtsson-Palme J, Kristiansson E, Larsson DGJ: Using metagenomics to investigate human and environmental resistomes – Methodological considerations. 8th Next Generation Sequencing Congress, London, Great Britain, 2016.
- Bengtsson-Palme J, Lundström SV, Flach C-F, Larsson DGJ: Determining the minimal selective concentrations of antibiotics in complex microbial communities. NDPIA Network Meeting 2016, Gothenburg, Sweden, 2016.
- Pal C, Bengtsson-Palme J, Kristiansson E, Larsson DGJ: Co-occurrence of resistance genes to antibiotics, biocides and metals on bacterial chromosomes and plasmids reveals novel insights into their co-selection potential. NDPIA Network Meeting 2016, Gothenburg, Sweden, 2016.
- Flach C-F, Svensson C-J, Pal C, Bengtsson-Palme J, Östman M, Kristiansson E, Larsson DGJ: Antifouling paint co-selects for antibiotic resistant bacteria in the marine environment. NDPIA Network Meeting 2016, Gothenburg, Sweden, 2016.
- Razavi M, Marathe NP, Bengtsson-Palme J, Flach C-F, Kristiansson E, Larsson DGJ: Identification of novel antibiotic resistance genes using amplicon sequencing of integrons from polluted environments in India. NDPIA Network Meeting 2016, Gothenburg, Sweden, 2016.
Conference talks
Presenting author indicated by *
- Pal C*, Bengtsson-Palme J, Kristiansson E, Larsson DGJ: Unravelling the structure and diversity of human, animal and environmental resistomes using metagenomics. NDPIA Network Meeting 2016, Gothenburg, Sweden, 2016.
- Bengtsson-Palme J*, Hammarén R, Pal C, Östman M, Björlenius B, Flach C-F, Kristiansson E, Fick J, Tysklind M, Larsson DGJ: Elucidating biocide and metal co-selection for antibiotic resistance in sewage treatment plants using metagenomics. The role of metals and biocides in the selection of antibiotic resistant bacteria, Stockholm, Sweden, 2016.
Conferences and workshops
- 8th Next Generation Sequencing Congress
in London, Great Britain (Poster) - NDPIA Network Meeting 2016
in Göteborg, Sweden (Poster) - The role of metals and biocides in the selection of antibiotic resistant bacteria
in Stockholm, Sweden (Talk)
Talks
- Antibiotic pollution from manufacturing
Antimicrobial resistance: the environmental impact, Healthcare Without Harm Webinar, 2016-11-03 - Elucidating biocide and metal co-selection for antibiotic resistance in sewage treatment plants using metagenomics
The role of metals and biocides in the selection of antibiotic resistant bacteria, Stockholm, Sweden, 2016-03-15
Research platforms and projects
- CARe
Centre for Antibiotic Resistance Research at University of Gothenburg - INTERACT
A FORMAS-funded project to investigate co-selection of metal, biocide and antibiotic resistance - UNITE
The database for DNA identification of fungi - GOTBIN – Gothenburg Bioinformatics Network
- SMAUG – Swedish Microbiome Analysis User Group
Other committees etc.
- Member of the expert panel for refinement of the criteria used for procurement of medicines in Sweden
- Member of the GOTBIN working group
- Reviewed manuscripts for Nature Reviews Microbiology, Bioinformatics, GigaScience, Proteomics Clinical Applications, Microbial Ecology, Molecular Ecology Resources, Nucleic Acids Research, Environmental Science and Technology, Water Research.