New beta brings major Metaxa2 updates
I am very happy to announce that a first public beta version of Metaxa2 version 2.2 has been released today! This new version brings two big and a number of small improvements to the Metaxa2 software (1). The first major addition is the introduction of the Metaxa2 Database Builder, which allows the user to create custom databases for virtually any genetic barcoding region. The second addition, which is related to the first, is that the classifier has been rewritten to have a more solid mathematical foundation. I have been promising that these updates were coming “soon” for one and a half years, but finally the end-product is good enough to see some real world testing. Bear in mind though that this is still a beta version that could contain obscure bugs. Here follows a list of new features (with further elaboration on a few below):
- The Metaxa2 Database Builder
- Support for additional barcoding genes, virtually any genetic region can now be used for taxonomic classification in Metaxa2
- The Metaxa2 database repository, which can be accessed through the new metaxa2_install_database tool
- Improved classification scoring model for better clarity and sensitivity
- A bundled COI database for athropods, showing off the capabilities of the database builder
- Support for compressed input files (gzip, zip, bzip, dsrc)
- Support for auto-detection of database locations
- Added output of probable taxonomic origin for sequences with reliability scores at each rank, made possible by the updated classifier
- Added the -x option for running only the extraction without the classification step
- Improved memory handling for very large rRNA datasets in the classifier (millions of sequences)
- This update also fixes a bug in the metaxa2_rf tool that could cause bias in very skewed datasets with small numbers of taxa
The new version of Metaxa2 can be downloaded here, and for those interested I will spend the rest of this post outlining the Metaxa2 Database Builder. The information below is also available in a slightly extended version in the software manual.
The major enhancement in Metaxa2 version 2.2 is the ability to use custom databases for classification. This means that the user can now make their own database for their own barcoding region of choice, or download additional databases from the Metaxa2 Database Repository. The selection of other databases is made through the “-g” option already existing in Metaxa2. As part of these changes, we have also updated the classification scoring model for better stringency and sensitivity across multiple databases and different genes. The old scoring system can still be used by specifying the –scoring_model option to “old”.
There are two different main operating modes of the Metaxa2 Database Builder, as well as a hybrid mode combining the features of the two other modes. The divergent and conserved modes work in almost completely different ways and deal with two different types of barcoding regions. The divergent mode is designed to deal with barcoding regions that exhibit fairly large variation between taxa within the same taxonomic domain. Such regions include, e.g., the eukaryotic ITS region, or the trnL gene used for plant barcoding. In the other mode – the conserved mode – a highly conserved barcoding region is expected (at least within the different taxonomic domains). Genes that fall into this category would be, e.g., the 16S SSU rRNA, and the bacterial rpoB gene. This option would most likely also be suitable for barcoding within certain groups of e.g. plants, where similarity of the barcoding regions can be expected to be high. There is also a third mode – the hybrid mode – that incorporates features of both the other. The hybrid mode is more experimental in nature, but could be useful in situations where both the other modes perform poorer than desired.
In the divergent (default) mode, the database builder will start by clustering the input sequences at 20% identity using USEARCH (2). All clusters generated from this process are then individually aligned using MAFFT (3). Those alignments are split into two regions, which are used to build two hidden Markov models for each cluster of sequences. These models will be less precise, but more sensitive than those generated in the conserved mode. In the divergent mode, the database builder will attempt to extract full-length sequences from the input data, but this may be less successful than in the conserved mode.
In the conserved mode, on the other hand, the database builder will first extract the barcoding region from the input sequences using models built from a reference sequence provided (see above) and the Metaxa2 extractor (1). It will then align all the extracted sequences using MAFFT and determine the conservation of each position in the alignment. When the criteria for degree of conservation are met, all conserved regions are extracted individually and are then re-aligned separately using MAFFT. The re-aligned sequences are used to build hidden Markov models representing the conserved regions with HMMER (4). In this mode, the classification database will only consist of the extracted full-length sequences.
In the hybrid mode, finally, the database builder will cluster the input sequences at 20% identity using USEARCH, and then proceed with the conserved mode approach on each cluster separately .
The actual taxonomic classification in Metaxa2 is done using a sequence database. It was shown in the original Metaxa2 paper that replacing the built-in database with a generic non-processed one was detrimental to performance in terms of accuracy (1). In the database builder, we have tried to incorporate some of the aspects of the manual database curation we did for the built-in database that can be automated. By default, all these filtration steps are turned off, but enabling them might drastically increase the accuracy of classifications based on the database.
To assess the accuracy of the constructed database, the Metaxa2 Database Builder allows for testing the detection ability and classification accuracy of the constructed database. This is done by sub-dividing the database sequences into subsets and rebuilding the database using a smaller (by default 90%), randomly selected, set of the sequence data (5). The remaining sequences (10% by default) are then classified using Metaxa2 with the subset database. The number of detections, and the numbers of correctly or incorrectly classified entries are recorded and averaged over a number of iterations (10 by default). This allows for obtaining a picture of the lower end of the accuracy of the database. However, since the evaluation only uses a subset of all sequences included in the full database, the performance of the full database actually constructed is likely to be slightly better. The evaluation can be turned on using the “–evaluate T” option.
Metaxa2 2.2 also introduces the database repository, from which the user can download additional databases for Metaxa2. To download new databases from the repository, the metaxa2_install_database command is used. This is a simple piece of software but requires internet access to function.
References
- Bengtsson-Palme J, Hartmann M, Eriksson KM, Pal C, Thorell K, Larsson DGJ, Nilsson RH: Metaxa2: Improved Identification and Taxonomic Classification of Small and Large Subunit rRNA in Metagenomic Data. Molecular Ecology Resources (2015). doi: 10.1111/1755-0998.12399 [Paper link]
- Edgar RC: Search and clustering orders of magnitude faster than BLAST. Bioinformatics, 26, 2460–2461 (2010).
- Katoh K, Standley DM: MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution, 30, 772–780 (2013).
- Eddy SR: Accelerated profile HMM searches. PLoS Computational Biology, 7, e1002195 (2011).
- Richardson RT, Bengtsson-Palme J, Johnson RM: Evaluating and Optimizing the Performance of Software Commonly Used for the Taxonomic Classification of DNA Sequence Data. Molecular Ecology Resources, 17, 4, 760–769 (2017). doi: 10.1111/1755-0998.12628
Published paper: Blastocystis and the intestinal microbiota
Yesterday, BMC Microbiology published a paper which I have co-authored with Joakim Forsell and his colleagues in at Umeå University. The paper (1) investigates the prevalence and subtype composition of Blastocystis – a eukaryotic microbe commonly present in the human intestine – among the 35 Swedish university students that we investigated for antibiotic resistance before and after travel to the Indian peninsula or Central Africa using shotgun metagenomics, and published in 2015 (2). In this paper, we used the same metagenomic data, but to assess the impact of travel on Blastocystis carriage and to understand the associations between Blastocystis and the bacterial gut microbiota. We found that 46% of the students carried Blastocystis before travel and 43% after. The two most commonly identified Blastocystis subtypes were ST3 and ST4, accounting for 20 of the 31 samples positive for Blastocystis. Interestingly, we detected no mixed subtype carriage in any individual, and all the ten individuals with a typable subtype before and after travel maintained their initial subtype.
Furthermore, we found that the composition of the gut bacterial community was not significantly altered between Blastocystis-carriers and non-carriers. Curiously, Blastocystis was accompanied with higher abundances of the bacterial genera Sporolactobacillus and Candidatus Carsonella. As perviously observed (3), Blastocystis carriage was positively associated with higher bacterial genus richness, and negatively correlated to the Bacteroides-driven enterotype. We, however, took this observation further, and could show that these associations were both largely driven by ST4 – a subtype commonly described in Europe – while the globally prevalent ST3 did not show such significant relationships.
The persistence of Blastocystis subtypes before and after travel indicates that long-term carriage of Blastocystis is common. The associations between Blastocystis and the bacterial microbiota found in this study could imply a link between Blastocystis and a healthy microbiota, as well as with diets high in vegetables. However, we cannot answer whether the associations between Blastocystis and the microbiota are resulting from the presence of Blastocystis per se, or are a prerequisite for colonization with Blastocystis, which are interesting opportunities for follow-up studies.
I think this type of data reuse for completely different questions is highly useful, and I am very happy that Joakim Forsell and his colleagues contacted me to hear if it was possible to do a Blastocystis screen of this data. The full paper can be read here.
References
- Forsell J, Bengtsson-Palme J, Angelin M, Johansson A, Evengård B, Granlund M: The relation between Blastocystis and the intestinal microbiota in Swedish travellers. BMC Microbiology, 17, 231 (2017). doi: 10.1186/s12866-017-1139-7 [Paper link]
- Bengtsson-Palme J, Angelin M, Huss M, Kjellqvist S, Kristiansson E, Palmgren H, Larsson DGJ, Johansson A: The human gut microbiome as a transporter of antibiotic resistance genes between continents. Antimicrobial Agents and Chemotherapy, 59, 10, 6551–6560 (2015). doi: 10.1128/AAC.00933-15 [Paper link]
- Andersen LO, Bonde I, Nielsen HB, Stensvold CR: A retrospective metagenomics approach to studying Blastocystis. FEMS Microbiology Ecology, 91, fiv072 (2015). doi: 10.1093/femsec/fiv072 [Paper link]
Published book chapter: Strategies for metagenomic analysis
Last summer, I was approached by Muniyandi Nagarajan to write a book chapter for a book on metagenomics. The book was published earlier this month, and is now available online (1). I have to admit that I have not yet read the entire book, but my own chapter deals with selecting the right tools for metagenomic analysis, and discusses different strategies to perform taxonomic classification, functional analysis, metagenomic assembly, and statistical comparisons between metagenomes (2). The chapter also considers the pros and cons of automated computational “pipelines” for analysis of metagenomic data. While I do not point to a specific set of software that obviously perform better in all situations, I do highlight some analysis strategies that clearly should be avoided. The chapter also suggests a few among the set of robust and well-functioning software tools that, in my opinion, should be used for metagenomic analyses. To some degree, this paper overlaps with the review paper we wrote on using metagenomics to analyze antibiotic resistance genes in various environments, published earlier this year (3), but the discussion in the book chapter is far more general. I imagine that the book chapter could be used, for example, in teaching metagenomics to students in bioinformatics (that’s at least a use I envision myself). Finally, apart from my own chapter, I can also highly recommend the chapter by Boulund et al. on statistical considerations for metagenomic data analysis (4). The book is available to buy from here, and the chapter can be read here.
References
- Nagarajan M (Ed.) Metagenomics: Perspectives, Methods, and Applications. ISBN: 9780081022689. Academic Press, Elsevier, USA (2018). doi: 10.1016/B978-0-08-102268-9 [Link]
- Bengtsson-Palme J: Strategies for Taxonomic and Functional Annotation of Metagenomes. In: Nagarajan M (Ed.) Metagenomics: Perspectives, Methods, and Applications, 55–79. Academic Press, Elsevier, USA (2018). doi: 10.1016/B978-0-08-102268-9.00003-3 [Link]
- Bengtsson-Palme J, Larsson DGJ, Kristiansson E: Using metagenomics to investigate human and environmental resistomes. Journal of Antimicrobial Chemotherapy, 72, 2690–2703 (2017). doi: 10.1093/jac/dkx199 [Paper link]
- Boulund F, Pereira MB, Jonsson V, Kristiansson E: Computational and Statistical Considerations in the Analysis of Metagenomic Data. In: Nagarajan M (Ed.) Metagenomics: Perspectives, Methods, and Applications, 81–102. Academic Press,, Elsevier, USA (2018). doi: 10.1016/B978-0-08-102268-9.00004-5 [Link]
Published opinion piece: Protection goals and risk assessment
Recently, Le Page et al. published a paper in Environmental International (1), partially building on the predicted no-effect concentrations for resistance selection for 111 antibiotics that me and Joakim Larsson published around two years ago (2). In their paper, the authors stress that discharge limits for antibiotics need to consider their potency to affect both environmental and human health, which we believe is a very reasonable standpoint, and to which we agree. However, we do not agree on the authors’ claim that cyanobacteria would often be more sensitive to antibiotics than the most sensitive human-associated bacteria (1). Importantly, we also think that it is a bit unclear from the paper which protection goals are considered. Are the authors mainly concerned with protecting microbial diversity in ecosystems, protecting ecosystem functions and services, or protecting from risks for resistance selection? This is important because it influence why one would want to mitigate, and therefore who would perform which actions. To elaborate a little on our standpoints, we wrote a short correspondence piece to Environment International, which is now published (3). (It has been online for a few days, but without a few last-minute changes we did to the proof, and hence I’m only posting about it now when the final version is online.) There is indeed an urgent need for discharge limits for antibiotics, particularly for industrial sources (4) and such limits would have tremendous value in regulation efforts, and in development of environmental criteria within public procurement and generic exchange programs (5). Importantly, while we are all for taking ecotoxicological data into account when doing risk assessment, we think that there should be solid scientific ground for mitigations and that regulations need to consider the benefits versus the costs, which is what we want to convey in our response to Le Page et al.
References
- Le Page G, Gunnarsson L, Snape J, Tyler CR: Integrating human and environmental health in antibiotic risk assessment: a critical analysis of protection goals, species sensitivity and antimicrobial resistance. Environment International, in press (2017). doi: 10.1016/j.envint.2017.09.013
- Bengtsson-Palme J, Larsson DGJ: Concentrations of antibiotics predicted to select for resistant bacteria: Proposed limits for environmental regulation. Environment International, 86, 140–149 (2016). doi: 10.1016/j.envint.2015.10.015
- Bengtsson-Palme J, Larsson DGJ: Protection goals must guide risk assessment for antibiotics. Environment International, in press (2017). doi: 10.1016/j.envint.2017.10.019
- Bengtsson-Palme J, Larsson DGJ: Time to limit antibiotic pollution. The Medicine Maker, 0416, 302, 17–18 (2016). [Paper link]
- Bengtsson-Palme J, Gunnarsson L, Larsson DGJ: Can branding and price of pharmaceuticals guide informed choices towards improved pollution control during manufacturing? Journal of Cleaner Production, 171, 137–146 (2018). doi: 10.1016/j.jclepro.2017.09.247
Published paper: Drug price is linked to environmental standards
Yesterday, Swedish television channel TV4 highlighted a recent publication by myself, Lina Gunnarsson and Joakim Larsson, in which we show that the price of pharmaceuticals is linked to the environmental standards of production countries. Surprisingly, however, this link seems to be mostly driven by whether the product is generic or original (branded), which in turns affect the prices.
In the study (1), published in Journal of Cleaner Production, we have used an exclusive set of Swedish sales data for pharmaceuticals combined with data on the origin of the active ingredients, obtained under an agreement to not identify individual manufacturers or products. We used this data to determine if price pressure and generic substitution could be linked to the general environmental performance and the corruption levels of the production countries, as measured by the Environmental Performance Index (2) and the Corruption Perception Index (3). In line with what we believed, India was the largest producer of generics, while Europe and the USA dominated the market for branded products (1). Importantly, we found that the price and environmental performance index of the production countries were linked, but that this relationship was largely explained by whether the product was original or generic.
To some extent, this relationship would allow buyers to select products that likely originate from countries that, in general terms, have better pollution control, which was also highlighted in the news clip that TV4 produced. However, what was lacking from that clip was the fact that this approach lacks resolution, because it does not say anything about the environmental footprint of individual products. We therefore conclude that to better allow consumers, hospitals and pharmacies to influence the environmental impact of their product choices, there is need for regulation and, importantly, transparency in the production chain, as has also been pointed out earlier (4,5). To this end, emissions from manufacturing need to be measured, allowing for control and follow-up on industry commitments towards sustainable manufacturing of pharmaceuticals (6). Since the discharges from pharmaceutical manufacturing not only leads to consequences to the local environment (7,8), but also in the case of antibiotics has potentially global consequences in terms of increasing risks for resistance development (9), limiting discharges is an urgent need to avoid a looming antibiotic resistance crisis (10).
The paper was also highlighted by the Centre for Antibiotic Resistance Research, and can be read here or here.
References
- Bengtsson-Palme J, Gunnarsson L, Larsson DGJ: Can branding and price of pharmaceuticals guide informed choices towards improved pollution control during manufacturing? Journal of Cleaner Production, 171, 137–146 (2018). doi: 10.1016/j.jclepro.2017.09.247
- Hsu A, Alexandre N, Cohen S, Jao P, Khusainova E: 2016 Environmental Performance Index. Yale University, New Haven, CT, USA (2016). http://epi.yale.edu/reports/2016-report
- Transparency International: Corruption Perceptions Index 2014. Transparency International, Berlin, Germany (2014). http://www.transparency.org/cpi2014/in_detail
- Larsson DGJ, Fick J: Transparency throughout the production chain–a way to reduce pollution from the manufacturing of pharmaceuticals? Regulatory Toxicology and Pharmacology, 53, 161–163 (2009). doi:10.1016/j.yrtph.2009.01.008
- Ågerstrand M, Berg C, Björlenius B, Breitholtz M, Brunström B, Fick J, Gunnarsson L, Larsson DGJ, Sumpter JP, Tysklind M, Rudén C: Improving environmental risk assessment of human pharmaceuticals. Environmental Science & Technology, 49, 5336–5345 (2015). doi:10.1021/acs.est.5b00302
- Industry Roadmap for Progress on Combating Antimicrobial Resistance: Industry Roadmap for Progress on Combating Antimicrobial Resistance – September 2016. (2016). http://www.ifpma.org/wp-content/uploads/2016/09/Roadmap-for-Progress-on-AMR-FINAL.pdf
- Larsson DGJ, de Pedro C, Paxeus N: Effluent from drug manufactures contains extremely high levels of pharmaceuticals. Journal of Hazardous Materials, 148, 751–755 (2007). doi:10.1016/j.jhazmat.2007.07.008
- aus der Beek T, Weber FA, Bergmann A, Hickmann S, Ebert I, Hein A, Küster A: Pharmaceuticals in the environment–Global occurrences and perspectives. Environmental Toxicology and Chemistry, 35, 823–835 (2016). doi:10.1002/etc.3339
- Bengtsson-Palme J, Larsson DGJ: Concentrations of antibiotics predicted to select for resistant bacteria: Proposed limits for environmental regulation. Environment International, 86, 140–149 (2016). doi: 10.1016/j.envint.2015.10.015
- Bengtsson-Palme J, Larsson DGJ: Time to limit antibiotic pollution. The Medicine Maker, 0416, 302, 17–18 (2016). [Paper link]
Published paper: Environmental factors leading to resistance
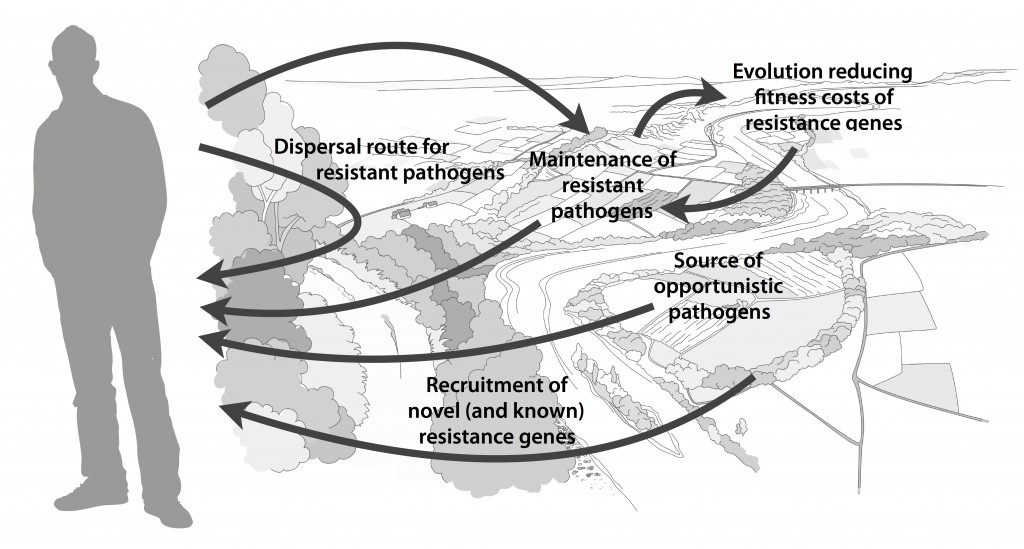
Myself, Joakim Larsson and Erik Kristiansson have written a review on the environmental factors that influence development and spread of antibiotic resistance, which was published today in FEMS Microbiology Reviews. The review (1) builds on thoughts developed in the latter parts of my PhD thesis (2), and seeks to provide a synthesis knowledge gained from different subfields towards the current understanding of evolutionary and ecological processes leading to clinical appearance of resistance genes, as well as the important environmental dispersal barriers preventing spread of resistant pathogens.
We postulate that emergence of novel resistance factors and mobilization of resistance genes are likely to occur continuously in the environment. However, the great majority of such genetic events are unlikely to lead to establishment of novel resistance factors in bacterial populations, unless there is a selection pressure for maintaining them or their fitness costs are negligible. To enable measures to prevent resistance development in the environment, it is therefore critical to investigate under what conditions and to what extent environmental selection for resistance takes place. Selection for resistance is likely less important for the dissemination of resistant bacteria, but will ultimately depend on how well the species or strain in question thrives in the external environment. Metacommunity theory (3,4) suggests that dispersal ability is central to this process, and therefore opportunistic pathogens with their main habitat in the environment may play an important role in the exchange of resistance factors between humans and the environment. Understanding the dispersal barriers hindering this exchange is not only key to evaluate risks, but also to prevent resistant pathogens, as well as novel resistance genes, from reaching humans.
Towards the end of the paper, we suggest certain environments that seem to be more important from a risk management perspective. We also discuss additional problems linked to the development of antibiotic resistance, such as increased evolvability of bacterial genomes (5) and which other types of genes that may be mobilized in the future, should the development continue (1,6). In this review, we also further develop thoughts on the relative risks of re-recruiting and spreading well-known resistance factors already circulating in pathogens, versus recruitment of completely novel resistance genes from environmental bacteria (7). While the latter case is likely to be very rare, and thus almost impossible to quantify the risks for, the consequences of such (potentially one-time) events can be dire.
I personally think that this is one of the best though-through pieces I have ever written, and since it is open access and (in my biased opinion) written in a fairly accessible way, I recommend everyone to read it. It builds on the ecological theories for resistance ecology developed by, among others, Fernando Baquero and José Martinez (8-13). Over the last year, it has been stressed several times at meetings (e.g. at the EDAR conferences in August) that there is a need to develop an ecological framework for antibiotic resistance genes. I think this paper could be one of the foundational pillars on such an endeavor and look forward to see how it will fit into the growing literature on the subject!
References
- Bengtsson-Palme J, Kristiansson E, Larsson DGJ: Environmental factors influencing the development and spread of antibiotic resistance. FEMS Microbiology Reviews, accepted manuscript (2017). doi: 10.1093/femsre/fux053
- Bengtsson-Palme J: Antibiotic resistance in the environment: a contribution from metagenomic studies. Doctoral thesis (medicine), Department of Infectious Diseases, Institute of Biomedicine, Sahlgrenska Academy, University of Gothenburg, 2016. [Link]
- Bengtsson J: Applied (meta)community ecology: diversity and ecosystem services at the intersection of local and regional processes. In: Verhoef HA, Morin PJ (eds.). Community Ecology: Processes, Models, and Applications. Oxford: Oxford University Press, 115–130 (2009).
- Leibold M, Norberg J: Biodiversity in metacommunities: Plankton as complex adaptive systems? Limnology and Oceanography, 1278–1289 (2004).
- Gillings MR, Stokes HW: Are humans increasing bacterial evolvability? Trends in Ecology and Evolution, 27, 346–352 (2012).
- Gillings MR: Evolutionary consequences of antibiotic use for the resistome, mobilome and microbial pangenome. Frontiers in Microbiology, 4, 4 (2013).
- Bengtsson-Palme J, Larsson DGJ: Antibiotic resistance genes in the environment: prioritizing risks. Nature Reviews Microbiology, 13, 369 (2015). doi: 10.1038/nrmicro3399-c1
- Baquero F, Alvarez-Ortega C, Martinez JL: Ecology and evolution of antibiotic resistance. Environmental Microbiology Reports, 1, 469–476 (2009).
- Baquero F, Tedim AP, Coque TM: Antibiotic resistance shaping multi-level population biology of bacteria. Frontiers in Microbiology, 4, 15 (2013).
- Berendonk TU, Manaia CM, Merlin C et al.: Tackling antibiotic resistance: the environmental framework. Nature Reviews Microbiology, 13, 310–317 (2015).
- Hiltunen T, Virta M, Laine A-L: Antibiotic resistance in the wild: an eco-evolutionary perspective. Philosophical Transactions of the Royal Society B: Biological Sciences, 372 (2017) doi: 10.1098/rstb.2016.0039.
- Martinez JL: Bottlenecks in the transferability of antibiotic resistance from natural ecosystems to human bacterial pathogens. Frontiers in Microbiology, 2, 265 (2011).
- Salyers AA, Amábile-Cuevas CF: Why are antibiotic resistance genes so resistant to elimination? Antimicrobial Agents and Chemotherapy, 41, 2321–2325 (1997).
Published paper: 76 new metallo-beta-lactamases
Today, Microbiome put online a paper lead-authored by my colleague Fanny Berglund – one of Erik Kristiansson‘s brilliant PhD students – in which we identify 76 novel metallo-ß-lactamases (1). This feat was made possible because of a new computational method designed by Fanny, which uses a hidden Markov model based on known B1 metallo-ß-lactamases. We analyzed over 10,000 bacterial genomes and plasmids and over 5 terabases of metagenomic data and could thereby predict 76 novel genes. These genes clustered into 59 new families of metallo-β-lactamases (given a 70% identity threshold). We also verified the functionality of 21 of these genes experimentally, and found that 18 were able to hydrolyze imipenem when inserted into Escherichia coli. Two of the novel genes contained atypical zinc-binding motifs in their active sites. Finally, we show that the B1 metallo-β-lactamases can be divided into five major groups based on their phylogenetic origin. It seems that nearly all of the previously characterized mobile B1 β-lactamases we identify in this study were likely to have originated from chromosomal genes present in species within the Proteobacteria, particularly Shewanella spp.
This study more than doubles the number of known B1 metallo-β-lactamases. As with the study by Boulund et al. (2) which we published last month on computational discovery of novel fluoroquinolone resistance genes (which used a very similar approach but on a completely different type of genes), this study also supports the hypothesis that environmental bacterial communities act as sources of uncharacterized antibiotic resistance genes (3-7). Fanny have done a fantastic job on this paper, and I highly recommend reading it in its entirety (it’s open access so you have virtually no excuse not to). It can be found here.
References
- Berglund F, Marathe NP, Österlund T, Bengtsson-Palme J, Kotsakis S, Flach C-F, Larsson DGJ, Kristiansson E: Identification of 76 novel B1 metallo-β-lactamases through large-scale screening of genomic and metagenomic data. Microbiome, 5, 134 (2017). doi: 10.1186/s40168-017-0353-8
- Boulund F, Berglund F, Flach C-F, Bengtsson-Palme J, Marathe NP, Larsson DGJ, Kristiansson E: Computational discovery and functional validation of novel fluoroquinolone resistance genes in public metagenomic data sets. BMC Genomics, 18, 682 (2017). doi: 10.1186/s12864-017-4064-0
- Bengtsson-Palme J, Larsson DGJ: Antibiotic resistance genes in the environment: prioritizing risks. Nature Reviews Microbiology, 13, 369 (2015). doi: 10.1038/nrmicro3399-c1
- Allen HK, Donato J, Wang HH et al.: Call of the wild: antibiotic resistance genes in natural environments. Nature Reviews Microbiology, 8, 251–259 (2010).
- Berendonk TU, Manaia CM, Merlin C et al.: Tackling antibiotic resistance: the environmental framework. Nature Reviews Microbiology, 13, 310–317 (2015).
- Martinez JL: Bottlenecks in the transferability of antibiotic resistance from natural ecosystems to human bacterial pathogens. Frontiers in Microbiology, 2, 265 (2011).
- Finley RL, Collignon P, Larsson DGJ et al.: The scourge of antibiotic resistance: the important role of the environment. Clinical Infectious Diseases, 57, 704–710 (2013).
ITSx in Bioconda
Mattias de Hollander at the Netherlands Institute of Ecology kindly informed me that they recently added the ITSx 1.1b version to the Bioconda package manager. This will make it easy for Conda users to install ITSx automatically into their systems and pipelines and also for others who are using conda. The Bioconda version can be found here. I would like to thank Mattias for this initiative and hope that the Bioconda version of ITSx will find much use!
Leaving Madison




Something to live up to




Around the biochemistry building of the University of Wisconsin-Madison campus (and perhaps also elsewhere), there are a lot of these signs, highlighting discoveries that were made in Madison. And they really have a history to be proud of. This place is where both bacterial conjugation and transduction was discovered, paving the way for much of the genome editing and studies we take for granted today. And they also did the basics required to generate knock-out mice for genetics studies. I could go on, but I I think I made my point. This place has a lost of history. And I doubt that my slightly messy project will live up to these groundbreaking discoveries.