Pfam + Wikipedia – finally!
Browsing the Pfam web site today, I discovered that the database finally has launched its Wikipedia co-ordination efforts.
This has happened along with the 25th release of the Pfam database (released 1st of April), and basically means that Wikipedia articles will be linked to Pfam families. Gradually, this will (hopefully) improve the annotation of Pfam families, which has in many cases been rather poor. The Xfam blog post related to Pfam release 25 says the change will be happening gradually, which might actually be good thing, given the quirks that might pop up.
(…) a major change is that Pfam annotation is now beginning to be co-ordinated via Wikipedia. Unlike Rfam, where every entry has a Wikipedia entry, we expect this to be a more gradual transition for Pfam, so not all entries currently have a corresponding Wikipedia article. For a more detailed discussion, check the help page. We actively encourage the addition of new/updated annotations via Wikipedia as they will appear far quicker than waiting for a Pfam release. If there are articles in Wikipedia that you think correspond to a family, then please mail us!
I have awaited this change for a long time, and is very happy that Pfam has finally taken this step. Congratulations and my sincerest thanks to the Pfam team! Now, let’s go editing!
Software reorganisation
I have reorganised my Software page a little bit, putting the smaller scripts on a separate page, to make the main software page tidier. The content of the pages is the same, and you still find bloutminer and metaorf on the main software page.
BLAST-mining software: bloutminer
I have put some “new” software online. I have had this piece of code lying around for some time but never got to upload it as I didn’t view it as “finished”. It is still not finished, but I would nevertheless like to share it with a wider audience. So, today I introduce bloutminer – the BLAST output mining script I have been using lately. bloutminer allows you to specify e.g. an E-value cutoff, a length cutoff and a percent identity cutoff, and extract a list of the hits satisfying these cutoffs. It takes table output (blastall option -m 8 ) as input. This is the software I used for the BLAST visualisation I have discussed earlier.
I normally use an E-value cutoff of 10 for my BLAST searches, and then extracts hits with bloutminer, allowing me to change the cutoffs at a later stage without redoing the whole BLAST search. You can also “pool” sequences into groups, based on their sequence tags. bloutminer is work in progress, and may contain nasty bugs. It can be found on the Software page. Please improve it at will.
Thesis presentation
I will present my master thesis “Metagenomic Analysis of Marine Periphyton Communities”, on Tuesday the 22nd of March, at 13.00. The presentation will take place in the room Folke Andreasson at Medicinaregatan 11 in Gothenburg. The presentation is open for everyone, but the number of seats are limited.
Underpinning Wikipedia’s Wisdom
In December, Alex Bateman, whose opinions on open science I support and have touched upon earlier, wrote a short correspondence letter to Nature [1] in which he again repeated the points of his talk at FEBS last summer. He concludes by the paragraph:
Many in the scientific community will admit to using Wikipedia occasionally, yet few have contributed content. For society’s sake, scientists must overcome their reluctance to embrace this resource.
I agree with this statement. However, as I also touched upon earlier, but like to repeat again – bold statements doesn’t make dreams come true – action does. Rfam, and the collaboration with RNA Biology and Wikipedia is a great example of such actions. So what other actions may be necessary to get researchers to contribute to the Wikipedian wisdom?
First of all, I do not think that the main obstacle to get researchers to edit Wikipedia articles is reluctance to doing so because Wikipedia is “inconsistent with traditional academic scholarship”, though that might be a partial explanation. What I think is the major problem is the time-reward tradeoff. Given the focus on publishing peer-reviewed articles, the race for higher impact factor, and the general tendency of measuring science by statistical measures, it should be no surprise that Wikipedia editing is far down on most scientists to-do lists, so also on mine. The reward of editing a Wikipedia article is a good feeling in your stomach that you have benefitted society. Good stomach feelings will, however, feed my children just as little as freedom of speech. Still, both Wikipedia editing and freedom of speech are extremely important, especially as a scientist.
Thus, there is a great need of a system that:
- Provides a reward or acknowledgement for Wikipedia editing.
- Makes Wikipedia editing economically sustainable.
- Encourages publishing of Wikipedia articles, or contributions to existing ones as part of the scientific publishing process.
Such a system could include a “contribution factor” similar to the impact factor, in which contribution of Wikipedia and other open access forums was weighted, with or without a usefulness measure. Such a usefulness measure could easily be determined by links from other Wikipedia articles, or similar. I realise that there would be severe drawbacks of such a system, similar to those of the impact factor system. I am not a huge fan of impact factors (read e.g. Per Seglen’s 1997 BMJ article [2] for some reasons why), but I do not see that system changing any time soon, and thus some kind of contribution factor could provide an additional statistical measure for evaluators to consider when examining scientists’ work.
While a contribution factor would be an incitement for researchers to contribute to the common knowledge, it will still not provide an economic value to do so. This could easily be changed by allowing, and maybe even requiring, scientists to contribute to Wikipedia and other public fora of scientific information as part of their science outreach duties. In fact, this public outreach duty (“tredje uppgiften” in Swedish) is governed in Swedish law. In 2009, the universities in Sweden have been assigned to “collaborate with the society and inform about their operations, and act such that scientific results produced at the university benefits society” (my translation). It seems rational that Wikipedia editing would be part of that duty, as that is the place were many (most?) people find information online today. Consequently, it is only up to the universities to demand 30 minutes of Wikipedia editing per week/month from their employees. Note here that I am referring to paid editing.
Another way of increasing the economic appeal of writing Wikipedia articles would be to encourage funding agencies and foundations to demand Wikipedia articles or similar as part of project reports. This would require researchers to make their findings public in order to get further funding, a move that would greatly increase the importance of increasing the common wisdom treasure. However, I suspect that many funding agencies, as well as researchers would be reluctant to such a solution.
Lastly, as shown by the Rfam/RNA Biology/Wikipedia relationship, scientific publishing itself could be tied to Wikipedia editing. This process could be started by e.g. open access journals such as PLoS ONE, either by demanding short Wikipedia notes to get an article published, or by simply provide prioritised publishing of articles which also have an accompanying Wiki-article. As mentioned previously, these short Wikipedia notes would also go through a peer-review process along with the full article. By tying this to the contribution factor, further incitements could be provided to get scientific progress in the hands of the general public.
Now, all these ideas put a huge burden on already hard-working scientists. I realise that they cannot all be introduced simultaneously. Opening up publishing requires time and thought, and should be done in small steps. But doing so is in the interest of scientists, the general public and the funders, as well as politicians. Because in the long run it will be hard to argue that society should pay for science when scientists are reluctant to even provide the public with an understandable version of the results. Instead of digging such a hole for ourselves, we should adapt the reward, evaluation, funding and publishing systems in a way that they benefit both researchers and the society we often say we serve.
- Bateman and Logan. Time to underpin Wikipedia wisdom. Nature (2010) vol. 468 (7325) pp. 765
- Seglen. Why the impact factor of journals should not be used for evaluating research. BMJ (1997) vol. 314 (7079) pp. 498-502
The research leaders of the future are… men
The Swedish Foundation for Strategic Research (SSF) has made public their grants to the research leaders of the future (link in Swedish), aiming to help and promote young researchers with a lot of potential and ambition to build their own research groups within their fields. 18 persons got 10 million SEK each (roughly 1.5 million USD), and also a leadership education. However, SSF obviously believes that men are superior in building and leading research groups, as 14 of the researchers were men (that’s 78%).
It is often argued that the reason that men get more and larger grants than women [1] is that they are more abundant in academia and that the over-representation of men will solve itself given sufficient time. This makes the SSF decisions particularly saddening. These 18 researchers represent the future of Swedish research, and SSF thinks that the research of the future is better of being led by… men. Alarmingly, the foundation’s statements on gender equality (in Swedish) says that (my translation):
The foundation for strategic research views gender equality as something self-evident, that should permeate not only the operations of the foundation, but also all activities that the foundation supports. Thus, the foundation strives towards that all treatment should be gender neutral, and that the under-represented gender should be given priority when other merits are similar. In an equal nation, research resources of men and women should always be taken advantage of, within all areas.
Still, only 20% of the chosen researchers are women. You may think this is a one-time-only event, but no, no, no, it’s much worse than this. In 2005, six of 18 researchers chosen were women (33%), in 2002 six out of 23 (26%), and 2008 six of 20 (30%). It seems that the SSF regards equality to mean 70% men, 30% women. That’s pretty bad for a foundation says it “views gender equality as something self-evident, that should permeate not only the operations of the foundation, but only all activities that the foundation supports.” Obviously, the words on equality are just words, and women still have a long way to go before treated equally by foundations supporting research.
In the long run, this inequality only cements the established norm with men on the top of the research departments. Wennerås and Wold wrote in 2000 that “junior scientists’ frustration at the pace of their scientific productivity is normal at the beginning of their careers, when they do most of the benchwork by themselves. But female scientists tend to remain at this level their entire working lives” [2]. Maybe it would be a good idea for the directors of the SSF to read this, and think about what their actions actually mean for the future of strategic research, and contemplate why women are leaving academia to a much larger extent than men [3]. Because research funders has a huge responsibility for the future of the scientific community.
References
- Wennerås and Wold. Nepotism and sexism in peer-review. Nature (1997) vol. 387 (6631) pp. 341-3
- Wennerås and Wold. A chair of one’s own. Nature (2000) vol. 408 (6813) pp. 647
- Handelsman et al. Careers in science. More women in science. Science (2005) vol. 309 (5738) pp. 1190-1
Assemblathon
There is currently an interesting competition going on organised by UCSC called the Assemblathon. The idea is that participating research groups will try to assemble simulated short-reads to a simulated genome, with the winner being the group doing it “best” (by some criteria set up by the evaluation team at the UC Davis Genome Center). The complete set of rules can be found here. The whole thing will culminate in a Genome Assembly Workshop at UC Santa Cruz in mid-March.
I think the competition is an interesting initiative, hopefully inspiring new, more efficient, sequence assembly ideas. Those are desperately needed in these times of ever-incresing DNA sequence generation. In addition, there are numerous already existing genome assembly programs, but (as noted on the Assemblathon site) it is not obvious which one is the best in a given situation. Hopefully the competition can shed some light on that too. The deadline for participation is the sixth of February, and even though I am not myself competent enough to participate, I hope the ones who do are successful in their work.
Raising the bar for genome sequencing
In a recent Nature article (1), Craig Venter and his co-workers at JCVI has not only sequenced one marine bacterium, but 137 different isolates. Their main goal of this study was to better understand the ecology of marine picoplankton in the context of Global Ocean Sampling (GOS) data (2,3). As I see it, there are at least two really interesting things going on here:
First, this is a milestone in sequencing. Were not talking one genome – one article anymore. Were talking one article – 137 new genomes. This vastly raises the bar for any sequencing efforts in the future, but even more importantly, it shifts the focus even further from the actual sequencing to the purpose of the sequencing. One sequenced genome might be interesting enough if it fills a biological knowledge gap, but just sequencing a bacterial strain isn’t worth that much anymore. With the arrival of second- and third-generation sequencing techniques, this development was pretty obvious, but this article is (to my knowledge) the first real proof of that this has finally happened. I expect that five to ten years from now, not sequencing an organism of interest for your research will be viewed as very strange and backwards-looking. “Why didn’t you sequence this?” will be a highly relevant review question for many publications. But also the days when you could write “we here publish for the first time the complete genome sequence of <insert organism name here>” and have that as the central theme for an article will soon be over. Sequencing will simply be reduced to the (valuable) tool it actually is. Which is probably good, as it brings us back to biology again. Articles like this one, where you look at ~200 genomes to investigate ecological questions, are simply providing a more relevant biological perspective than staring at the sequence of one genome in a time when DNA-data is flooding over us.
Second, this is the first (again, to my knowledge) publication where questions arising from metagenomics (2,3,4) has initiated a huge sequencing effort to understand the ecology or the environment to which the metagenome is associated. This highlights a new use of metagenomics as a prospective technique, to mine various environments for interesting features, and then select a few of its inhabitants and look closer at who is responsible for what. With a number of emerging single cell sequencing and visualisation techniques (5,6,7,8) as well as the application of cell sorting approaches to environmental communities (5,9), we can expect metagenomics to play a huge role in organism, strain and protein discovery, but also in determining microbial ecosystem services. Though Venter’s latest article (1) is just a first step towards this new role for metagenomics, it’s a nice example of what (meta)genomics could look like towards the end of this decade, if even not sooner.
- Yooseph et al. Genomic and functional adaptation in surface ocean planktonic prokaryotes. Nature (2010) vol. 468 (7320) pp. 60-6
- Yooseph et al. The Sorcerer II Global Ocean Sampling expedition: expanding the universe of protein families. Plos Biol (2007) vol. 5 (3) pp. e16
- Rusch et al. The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. Plos Biol (2007) vol. 5 (3) pp. e77
- Rusch et al. Characterization of Prochlorococcus clades from iron-depleted oceanic regions. Proceedings of the National Academy of Sciences of the United States of America (2010) pp.
- Woyke et al. Assembling the marine metagenome, one cell at a time. PLoS ONE (2009) vol. 4 (4) pp. e5299
- Woyke et al. One bacterial cell, one complete genome. PLoS ONE (2010) vol. 5 (4) pp. e10314
- Moraru et al. GeneFISH – an in situ technique for linking gene presence and cell identity in environmental microorganisms. Environ Microbiol (2010) pp.
- Lasken. Genomic DNA amplification by the multiple displacement amplification (MDA) method. Biochem Soc Trans (2009) vol. 37 (Pt 2) pp. 450-3
- Mary et al. Metaproteomic and metagenomic analyses of defined oceanic microbial populations using microwave cell fixation and flow cytometric sorting. FEMS microbiology ecology (2010) pp.
Alex Bateman says protein family wiki is coming – but when?
Nature recently had a nice news article on Bio-wikis and biological databases connected to Wikipedia where Alex Bateman says they’re working on a protein-family wiki that will be hosted on Wikipedia, similar to the Rfam wiki, which he talked about at FEBS this summer. I am of course very excited about this, and hope that the new Pfam (?) wiki will come rather sooner than later. As pointed out earlier, the Nature article also underlines the problem of scientific wikis; currently there is no career incentive to get researchers to spend their time editing wiki-articles. That is a shame, and perhaps a system copying the system of Rfam and RNA Biology could help in this direction. The only question is which journal(s) that would be interested in such a commitment to open science…
BLAST Visualization
Perhaps because of my roots in systems biology (or the cause of going there in the first place), I have always had an interest in creating visually appealing images of data, many times in the form of networks. I find that often in bioinformatics, one of the hardest problems is to make information understandable. For example, a BLAST output might say very little about how the genes or proteins are connected to each other, at least to the untrained eye.
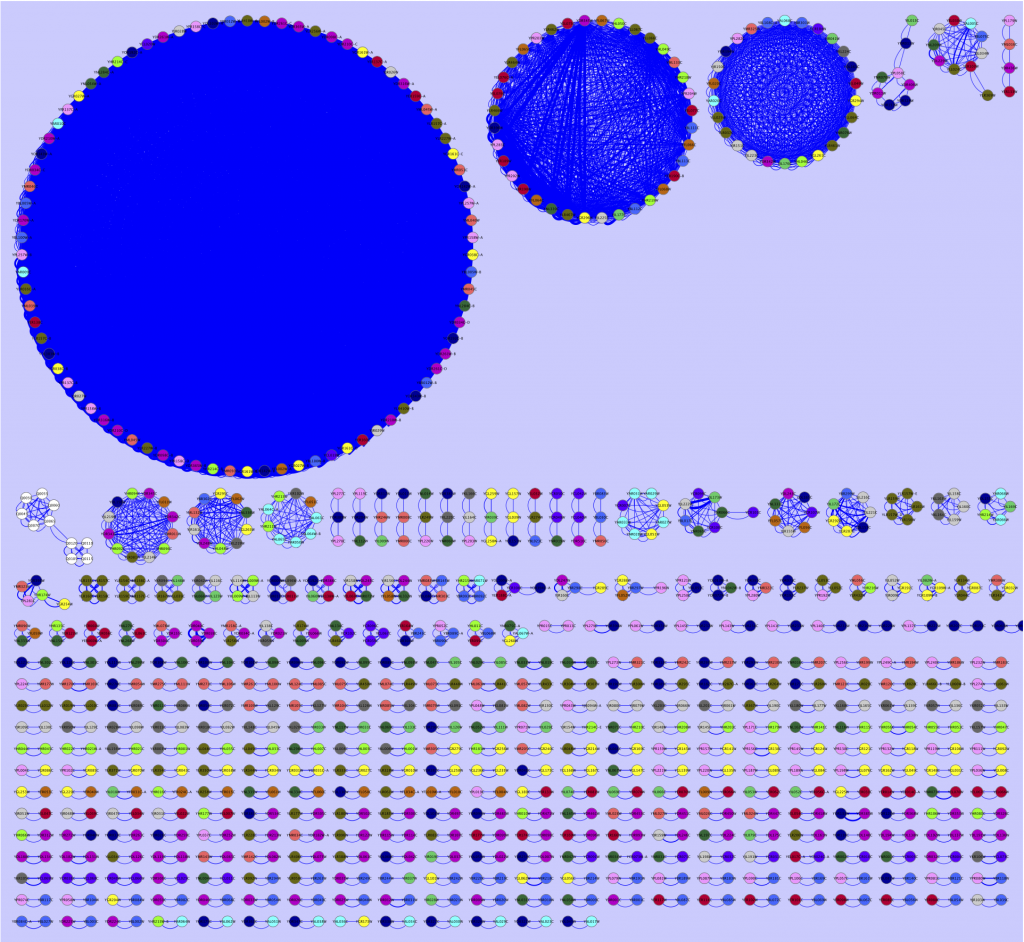
Therefore, during the last weeks I have fiddled around with various ways of viewing interesting portions of BLAST reports. By making all-against-all BLAST searches, and outputting the data in table format (blastall option -m 8), I have been able to extract the hits I am interested in and export them into a Cytoscape compatible format, with some accompanying metadata (scores, e-values, alignment length, etc.). The results are many times pretty unparsable by the eye, rendering them a bit meaningless, but have been more and more interesting as I have put more effort into the extraction script. Just as an example, I here provide a simple map of the best all-against-all matches in the Saccharomyces cerevisiae genome, as a Cytoscape network (click for full size):
The largest circle consists of transposable elements (jumping DNA which inserts itself at multiple locations in the genome, no surprise there is a lot of them, and that these are pretty conserved). The circle to the left of the transposon circle consists of genes located inside the telomeric regions. Why they show such high similarity I do not know, but it seems plausible that the telomere thing could play a role here. The third circle contain mostly members of the seripauperin multigene family, which is also located close to the telomeres. At the bottom you found the gene pairs, that match to each other. You could go on with all the smaller structures as well, but I am no yeast expert, so I will stop here, letting this serve as an example of what a BLAST report really look like.
For this image, I have used a blastn report of all yeast ORFs (taken from yeastgenome.org) as input to my extraction tool, selected Cytoscape compatible output, and used a maximal e-value of 0.00001 and an alignment length of at least 50 nts as criteria to be extracted. I have also pooled the sequences according to chromosome number. The pooling was used to color code the nodes in Cytoscape. The edge width is connected to alignment score, a high score renders a thick line, and a low score causes the line to be thin.
I am still working on the extraction tool and will not provide any code yet. Input would, however, be appreciated. My personal opinion is that in the near future, the overload of newly produced DNA and protein sequences will choke us if do not come up with more intuitive ways of displaying data. I don’t think that the network above is there yet. Still, it conveys information I would not have been able to understand from just looking at the BLAST output. The first attempts to come around the sequence overload problem won’t be the best ones. But we got to start working on visualization methods today, so that we do not end up with sequences over our shoulders in just a few years. Besides, a network image seems much more impressive than a number of lines of text…