New beta brings major Metaxa2 updates
I am very happy to announce that a first public beta version of Metaxa2 version 2.2 has been released today! This new version brings two big and a number of small improvements to the Metaxa2 software (1). The first major addition is the introduction of the Metaxa2 Database Builder, which allows the user to create custom databases for virtually any genetic barcoding region. The second addition, which is related to the first, is that the classifier has been rewritten to have a more solid mathematical foundation. I have been promising that these updates were coming “soon” for one and a half years, but finally the end-product is good enough to see some real world testing. Bear in mind though that this is still a beta version that could contain obscure bugs. Here follows a list of new features (with further elaboration on a few below):
- The Metaxa2 Database Builder
- Support for additional barcoding genes, virtually any genetic region can now be used for taxonomic classification in Metaxa2
- The Metaxa2 database repository, which can be accessed through the new metaxa2_install_database tool
- Improved classification scoring model for better clarity and sensitivity
- A bundled COI database for athropods, showing off the capabilities of the database builder
- Support for compressed input files (gzip, zip, bzip, dsrc)
- Support for auto-detection of database locations
- Added output of probable taxonomic origin for sequences with reliability scores at each rank, made possible by the updated classifier
- Added the -x option for running only the extraction without the classification step
- Improved memory handling for very large rRNA datasets in the classifier (millions of sequences)
- This update also fixes a bug in the metaxa2_rf tool that could cause bias in very skewed datasets with small numbers of taxa
The new version of Metaxa2 can be downloaded here, and for those interested I will spend the rest of this post outlining the Metaxa2 Database Builder. The information below is also available in a slightly extended version in the software manual.
The major enhancement in Metaxa2 version 2.2 is the ability to use custom databases for classification. This means that the user can now make their own database for their own barcoding region of choice, or download additional databases from the Metaxa2 Database Repository. The selection of other databases is made through the “-g” option already existing in Metaxa2. As part of these changes, we have also updated the classification scoring model for better stringency and sensitivity across multiple databases and different genes. The old scoring system can still be used by specifying the –scoring_model option to “old”.
There are two different main operating modes of the Metaxa2 Database Builder, as well as a hybrid mode combining the features of the two other modes. The divergent and conserved modes work in almost completely different ways and deal with two different types of barcoding regions. The divergent mode is designed to deal with barcoding regions that exhibit fairly large variation between taxa within the same taxonomic domain. Such regions include, e.g., the eukaryotic ITS region, or the trnL gene used for plant barcoding. In the other mode – the conserved mode – a highly conserved barcoding region is expected (at least within the different taxonomic domains). Genes that fall into this category would be, e.g., the 16S SSU rRNA, and the bacterial rpoB gene. This option would most likely also be suitable for barcoding within certain groups of e.g. plants, where similarity of the barcoding regions can be expected to be high. There is also a third mode – the hybrid mode – that incorporates features of both the other. The hybrid mode is more experimental in nature, but could be useful in situations where both the other modes perform poorer than desired.
In the divergent (default) mode, the database builder will start by clustering the input sequences at 20% identity using USEARCH (2). All clusters generated from this process are then individually aligned using MAFFT (3). Those alignments are split into two regions, which are used to build two hidden Markov models for each cluster of sequences. These models will be less precise, but more sensitive than those generated in the conserved mode. In the divergent mode, the database builder will attempt to extract full-length sequences from the input data, but this may be less successful than in the conserved mode.
In the conserved mode, on the other hand, the database builder will first extract the barcoding region from the input sequences using models built from a reference sequence provided (see above) and the Metaxa2 extractor (1). It will then align all the extracted sequences using MAFFT and determine the conservation of each position in the alignment. When the criteria for degree of conservation are met, all conserved regions are extracted individually and are then re-aligned separately using MAFFT. The re-aligned sequences are used to build hidden Markov models representing the conserved regions with HMMER (4). In this mode, the classification database will only consist of the extracted full-length sequences.
In the hybrid mode, finally, the database builder will cluster the input sequences at 20% identity using USEARCH, and then proceed with the conserved mode approach on each cluster separately .
The actual taxonomic classification in Metaxa2 is done using a sequence database. It was shown in the original Metaxa2 paper that replacing the built-in database with a generic non-processed one was detrimental to performance in terms of accuracy (1). In the database builder, we have tried to incorporate some of the aspects of the manual database curation we did for the built-in database that can be automated. By default, all these filtration steps are turned off, but enabling them might drastically increase the accuracy of classifications based on the database.
To assess the accuracy of the constructed database, the Metaxa2 Database Builder allows for testing the detection ability and classification accuracy of the constructed database. This is done by sub-dividing the database sequences into subsets and rebuilding the database using a smaller (by default 90%), randomly selected, set of the sequence data (5). The remaining sequences (10% by default) are then classified using Metaxa2 with the subset database. The number of detections, and the numbers of correctly or incorrectly classified entries are recorded and averaged over a number of iterations (10 by default). This allows for obtaining a picture of the lower end of the accuracy of the database. However, since the evaluation only uses a subset of all sequences included in the full database, the performance of the full database actually constructed is likely to be slightly better. The evaluation can be turned on using the “–evaluate T” option.
Metaxa2 2.2 also introduces the database repository, from which the user can download additional databases for Metaxa2. To download new databases from the repository, the metaxa2_install_database command is used. This is a simple piece of software but requires internet access to function.
References
- Bengtsson-Palme J, Hartmann M, Eriksson KM, Pal C, Thorell K, Larsson DGJ, Nilsson RH: Metaxa2: Improved Identification and Taxonomic Classification of Small and Large Subunit rRNA in Metagenomic Data. Molecular Ecology Resources (2015). doi: 10.1111/1755-0998.12399 [Paper link]
- Edgar RC: Search and clustering orders of magnitude faster than BLAST. Bioinformatics, 26, 2460–2461 (2010).
- Katoh K, Standley DM: MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution, 30, 772–780 (2013).
- Eddy SR: Accelerated profile HMM searches. PLoS Computational Biology, 7, e1002195 (2011).
- Richardson RT, Bengtsson-Palme J, Johnson RM: Evaluating and Optimizing the Performance of Software Commonly Used for the Taxonomic Classification of DNA Sequence Data. Molecular Ecology Resources, 17, 4, 760–769 (2017). doi: 10.1111/1755-0998.12628
Published book chapter: Strategies for metagenomic analysis
Last summer, I was approached by Muniyandi Nagarajan to write a book chapter for a book on metagenomics. The book was published earlier this month, and is now available online (1). I have to admit that I have not yet read the entire book, but my own chapter deals with selecting the right tools for metagenomic analysis, and discusses different strategies to perform taxonomic classification, functional analysis, metagenomic assembly, and statistical comparisons between metagenomes (2). The chapter also considers the pros and cons of automated computational “pipelines” for analysis of metagenomic data. While I do not point to a specific set of software that obviously perform better in all situations, I do highlight some analysis strategies that clearly should be avoided. The chapter also suggests a few among the set of robust and well-functioning software tools that, in my opinion, should be used for metagenomic analyses. To some degree, this paper overlaps with the review paper we wrote on using metagenomics to analyze antibiotic resistance genes in various environments, published earlier this year (3), but the discussion in the book chapter is far more general. I imagine that the book chapter could be used, for example, in teaching metagenomics to students in bioinformatics (that’s at least a use I envision myself). Finally, apart from my own chapter, I can also highly recommend the chapter by Boulund et al. on statistical considerations for metagenomic data analysis (4). The book is available to buy from here, and the chapter can be read here.
References
- Nagarajan M (Ed.) Metagenomics: Perspectives, Methods, and Applications. ISBN: 9780081022689. Academic Press, Elsevier, USA (2018). doi: 10.1016/B978-0-08-102268-9 [Link]
- Bengtsson-Palme J: Strategies for Taxonomic and Functional Annotation of Metagenomes. In: Nagarajan M (Ed.) Metagenomics: Perspectives, Methods, and Applications, 55–79. Academic Press, Elsevier, USA (2018). doi: 10.1016/B978-0-08-102268-9.00003-3 [Link]
- Bengtsson-Palme J, Larsson DGJ, Kristiansson E: Using metagenomics to investigate human and environmental resistomes. Journal of Antimicrobial Chemotherapy, 72, 2690–2703 (2017). doi: 10.1093/jac/dkx199 [Paper link]
- Boulund F, Pereira MB, Jonsson V, Kristiansson E: Computational and Statistical Considerations in the Analysis of Metagenomic Data. In: Nagarajan M (Ed.) Metagenomics: Perspectives, Methods, and Applications, 81–102. Academic Press,, Elsevier, USA (2018). doi: 10.1016/B978-0-08-102268-9.00004-5 [Link]
Published paper: 76 new metallo-beta-lactamases
Today, Microbiome put online a paper lead-authored by my colleague Fanny Berglund – one of Erik Kristiansson‘s brilliant PhD students – in which we identify 76 novel metallo-ß-lactamases (1). This feat was made possible because of a new computational method designed by Fanny, which uses a hidden Markov model based on known B1 metallo-ß-lactamases. We analyzed over 10,000 bacterial genomes and plasmids and over 5 terabases of metagenomic data and could thereby predict 76 novel genes. These genes clustered into 59 new families of metallo-β-lactamases (given a 70% identity threshold). We also verified the functionality of 21 of these genes experimentally, and found that 18 were able to hydrolyze imipenem when inserted into Escherichia coli. Two of the novel genes contained atypical zinc-binding motifs in their active sites. Finally, we show that the B1 metallo-β-lactamases can be divided into five major groups based on their phylogenetic origin. It seems that nearly all of the previously characterized mobile B1 β-lactamases we identify in this study were likely to have originated from chromosomal genes present in species within the Proteobacteria, particularly Shewanella spp.
This study more than doubles the number of known B1 metallo-β-lactamases. As with the study by Boulund et al. (2) which we published last month on computational discovery of novel fluoroquinolone resistance genes (which used a very similar approach but on a completely different type of genes), this study also supports the hypothesis that environmental bacterial communities act as sources of uncharacterized antibiotic resistance genes (3-7). Fanny have done a fantastic job on this paper, and I highly recommend reading it in its entirety (it’s open access so you have virtually no excuse not to). It can be found here.
References
- Berglund F, Marathe NP, Österlund T, Bengtsson-Palme J, Kotsakis S, Flach C-F, Larsson DGJ, Kristiansson E: Identification of 76 novel B1 metallo-β-lactamases through large-scale screening of genomic and metagenomic data. Microbiome, 5, 134 (2017). doi: 10.1186/s40168-017-0353-8
- Boulund F, Berglund F, Flach C-F, Bengtsson-Palme J, Marathe NP, Larsson DGJ, Kristiansson E: Computational discovery and functional validation of novel fluoroquinolone resistance genes in public metagenomic data sets. BMC Genomics, 18, 682 (2017). doi: 10.1186/s12864-017-4064-0
- Bengtsson-Palme J, Larsson DGJ: Antibiotic resistance genes in the environment: prioritizing risks. Nature Reviews Microbiology, 13, 369 (2015). doi: 10.1038/nrmicro3399-c1
- Allen HK, Donato J, Wang HH et al.: Call of the wild: antibiotic resistance genes in natural environments. Nature Reviews Microbiology, 8, 251–259 (2010).
- Berendonk TU, Manaia CM, Merlin C et al.: Tackling antibiotic resistance: the environmental framework. Nature Reviews Microbiology, 13, 310–317 (2015).
- Martinez JL: Bottlenecks in the transferability of antibiotic resistance from natural ecosystems to human bacterial pathogens. Frontiers in Microbiology, 2, 265 (2011).
- Finley RL, Collignon P, Larsson DGJ et al.: The scourge of antibiotic resistance: the important role of the environment. Clinical Infectious Diseases, 57, 704–710 (2013).
ITSx in Bioconda
Mattias de Hollander at the Netherlands Institute of Ecology kindly informed me that they recently added the ITSx 1.1b version to the Bioconda package manager. This will make it easy for Conda users to install ITSx automatically into their systems and pipelines and also for others who are using conda. The Bioconda version can be found here. I would like to thank Mattias for this initiative and hope that the Bioconda version of ITSx will find much use!
Published paper: Computational discovery of novel qnr genes
BMC Genomics today published a paper first-authored by my long-time colleague Fredrik Boulund, which describes a computational screen of genomes and metagenomes for novel qnr fluoroquinolone resistance genes (1). The study makes use of Fredrik’s well-designed and updated qnr-prediction pipeline, but in contrast to his previous publication based on the pipeline from 2012 (2), we here study a 20-fold larger dataset of almost 13 terabases of sequence data. Based on this data, the pipeline predicted 611 putative qnr genes, including all previously described plasmid-mediated qnr gene families. 20 of the predicted genes were previously undescribed, and of these nine were selected for experimental validation. Six of those tested genes improved the survivability under ciprofloxacin exposure when expressed in Escherichia coli. The study shows that qnr genes are almost ubiquitous in environmental microbial communities. This study also lends further credibility to the hypothesis that environmental bacterial communities can act as sources of previously uncharacterized antibiotic resistance genes (3-7). The study can be read in its entirety here.
References
- Boulund F, Berglund F, Flach C-F, Bengtsson-Palme J, Marathe NP, Larsson DGJ, Kristiansson E: Computational discovery and functional validation of novel fluoroquinolone resistance genes in public metagenomic data sets. BMC Genomics, 18, 682 (2017). doi: 10.1186/s12864-017-4064-0
- Boulund F, Johnning A, Pereira MB, Larsson DGJ, Kristiansson E: A novel method to discover fluoroquinolone antibiotic resistance (qnr) genes in fragmented nucleotide sequences. BMC Genomics, 13, 695 (2012). doi: 10.1186/1471-2164-13-695
- Bengtsson-Palme J, Larsson DGJ: Antibiotic resistance genes in the environment: prioritizing risks. Nature Reviews Microbiology, 13, 369 (2015). doi: 10.1038/nrmicro3399-c1
- Allen HK, Donato J, Wang HH et al.: Call of the wild: antibiotic resistance genes in natural environments. Nature Reviews Microbiology, 8, 251–259 (2010).
- Berendonk TU, Manaia CM, Merlin C et al.: Tackling antibiotic resistance: the environmental framework. Nature Reviews Microbiology, 13, 310–317 (2015).
- Martinez JL: Bottlenecks in the transferability of antibiotic resistance from natural ecosystems to human bacterial pathogens. Frontiers in Microbiology, 2, 265 (2011).
- Finley RL, Collignon P, Larsson DGJ et al.: The scourge of antibiotic resistance: the important role of the environment. Clinical Infectious Diseases, 57, 704–710 (2013).
Published paper: The Calanus glacialis mitogenome
Mitochondrial DNA Part B today published a mitochondrial genome announcement paper (1) in which I was involved in doing the assemblies and annotating them. The paper describes the mitogenome of Calanus glacialis, a marine planktonic copepod, which is a keystone species in the Arctic Ocean. The mitogenome is 20,674 bp long, and includes 13 protein-coding genes, 2 rRNA genes and 22 tRNA genes. While this is of course note a huge paper, we believe that this new resource will be of interest in understanding the structure and dynamics of C. glacialis populations. The main work in this paper has been carried out by Marvin Choquet at Nord University in Bodø, Norway. So hats off to him for great work, thanks Marvin! The paper can be read here.
Reference
- Choquet M, Alves Monteiro HJ, Bengtsson-Palme J, Hoarau G: The complete mitochondrial genome of the copepod Calanus glacialis. Mitochondrial DNA Part B, 2, 2, 506–507 (2017). doi: 10.1080/23802359.2017.1361357 [Paper link]
Published paper: Helicobacter pylori vs. the viable gastric microbiota
I have just returned from a week of vacation in Sicily (almost without internet access), so I am a tad late to this news, but earlier this week Infection and Immunity published our paper on the Helicobacter pylori transcriptome in gastric infection (and early stages of carcinogenesis), and how that relates to the transcriptionally active microbiota in the stomach environment (1). This paper has been long in the making (an earlier version of it was included in Kaisa Thorell’s PhD thesis (2)), but some late additional analyses did substantially strengthen our confidence in the suggestions we got from the original data.
In the paper (1) we use metatranscriptomic RNAseq to investigate the composition of the viable microbial community, and at the same time study H. pylori gene expression in stomach biopsies. The biopsies were sampled from individuals with different degrees of H. pylori infection and/or pre-malignant tissue changes. We found that H. pylori completely dominates the microbiota in infected individuals, but (somewhat surprisingly) also in the majority of individuals classified as H. pylori uninfected using traditional methods. This confirms previous reports that have detected minute quantities of H. pylori also in presumably uninfected individuals (3-6), and raises the question of how large part of the human population (if any) that is truly not infected/colonized by H. pylori. The abundance of H. pylori was correlated with the abundance of Campylobacter, Deinococcus, and Sulfurospirillum. It is unclear, however, if these genera only share the same habitat preferences as Helicobacter, or if they are specifically promoted by the presence of H. pylori (or tissue changes induced by it). We also found that genes involved in pH regulation and nickel transport were highly expressed in H. pylori, regardless of disease stage. As far as we know, this study is the first to use metatranscriptomics to study the viable microbiota of the human stomach, and we think that this is a promising approach for future studies on pathogen-microbiota interactions. The paper (in unedited format) can be read here.
References
- Thorell K, Bengtsson-Palme J, Liu OH, Gonzales RVP, Nookaew I, Rabeneck L, Paszat L, Graham DY, Nielsen J, Lundin SB, Sjöling Å: In vivo analysis of the viable microbiota and Helicobacter pylori transcriptome in gastric infection and early stages of carcinogenesis. Infection and Immunity, accepted manuscript (2017). doi: 10.1128/IAI.00031-17 [Paper link]
- Thorell K: Multi-level characterization of host and pathogen in Helicobacter pylori-associated gastric carcinogenesis. Doctoral thesis, Institute of Biomedicine, Sahlgrenska Academy, University of Gothenburg (2014). [Link]
- Bik EM, Eckburg PB, Gill SR, Nelson KE, Purdom EA, Francois F, Perez-Perez G, Blaser MJ, Reman DA: Molecular analysis of the bacterial microbiota in the human stomach. PNAS, 103:732-737 (2006).
- Dicksved J, Lindberg M, Rosenquist M, Enroth H, Jansson JK, Engstrand L: Molecular characterization of the stomach microbiota in patients with gastric cancer and in controls. Journal of Medical Microbiology, 58:509-516 (2009).
- Maldonado-Contreras A, Goldfarb KC, Godoy-Vitorino F, Karaoz U, Contreras M, Blaser MJ, Brodie EL, Dominguez-Bello MG: Structure of the human gastric bacterial community in relation to Helicobacter pylori status. ISME Journal, 5:574-579 (2011).
- Li TH, Qin Y, Sham PC, Lau KS, Chu KM, Leung WK: Alterations in Gastric Microbiota After H. Pylori Eradication and in Different Histological Stages of Gastric Carcinogenesis. Scientific Reports, 7:44935 (2017).
Major ITSx update (beta version)
Today, I am very happy to announce that after years in the making and months in testing, the next generation of ITSx, version 1.1, is ready to step into the public light and scrutiny. I have today uploaded a public beta version of the ITSx 1.1 release, which I encourage everyone that have enjoyed using ITSx to try out.
The 1.1 release of ITSx includes a wide range of new feature, including:
- A 2-10x performance increase (depending on the dataset), since ITSx now utilizes hmmsearch instead of hmmscan to detect the ITS regions and distributes the CPU cores better
- Improved ITS detection among fungi and chlorophyta, by addition of new HMM-profiles
- The HMM profile format for ITSx has been updated to HMMER3/f (thus ITSx now requires HMMER version 3.1 or later)
- Better handling of interrupted HMMER searches
- Added the
--require_anchoroption to only include sequences where the complete anchor is found in the output - Added the possibility for partial sequence output for the SSU, LSU and 5.8S regions
- Fixed a bug causing problems when reading sequence data from standard input
A lot of the code has changed in this version, which means that there might still be bugs lingering in the program. Since I will be on vacation throughout July, I encourage everyone to submit bug reports and questions, but I will not promise to respond to them until in August.
I hope that you will enjoy this new ITSx release, which you can download here. Happy barcoding!
Published paper: Investigating resistomes using metagenomics
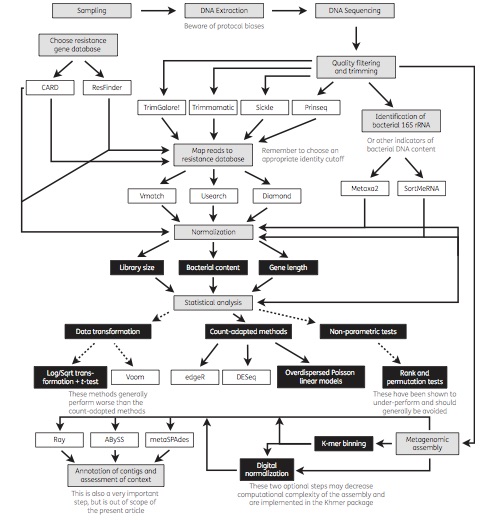
Today, a review paper which I wrote together with Joakim Larsson and Erik Kristiansson was published in Journal of Antimicrobial Chemotherapy (1). We have for a long time used metagenomic DNA sequencing to study antibiotic resistance in different environments (2-6), including in the human microbiota (7). Generally, our ultimate purpose has been to assess the risks to human health associated with resistance genes in the environment. However, a multitude of methods exist for metagenomic data analysis, and over the years we have learned that not all methods are suitable for the investigation of resistance genes for this purpose. In our review paper, we describe and discuss current methods for sequence handling, mapping to databases of resistance genes, statistical analysis and metagenomic assembly. We also provide an overview of important considerations related to the analysis of resistance genes, and end by recommending some of the currently used tools, databases and methods that are best equipped to inform research and clinical practice related to antibiotic resistance (see the figure from the paper below). We hope that the paper will be useful to researchers and clinicians interested in using metagenomic sequencing to better understand the resistance genes present in environmental and human-associated microbial communities.
References
- Bengtsson-Palme J, Larsson DGJ, Kristiansson E: Using metagenomics to investigate human and environmental resistomes. Journal of Antimicrobial Chemotherapy, advance access (2017). doi: 10.1093/jac/dkx199 [Paper link]
- Bengtsson-Palme J, Boulund F, Fick J, Kristiansson E, Larsson DGJ: Shotgun metagenomics reveals a wide array of antibiotic resistance genes and mobile elements in a polluted lake in India. Frontiers in Microbiology, 5, 648 (2014). doi: 10.3389/fmicb.2014.00648 [Paper link]
- Lundström S, Östman M, Bengtsson-Palme J, Rutgersson C, Thoudal M, Sircar T, Blanck H, Eriksson KM, Tysklind M, Flach C-F, Larsson DGJ: Minimal selective concentrations of tetracycline in complex aquatic bacterial biofilms. Science of the Total Environment, 553, 587–595 (2016). doi: 10.1016/j.scitotenv.2016.02.103 [Paper link]
- Bengtsson-Palme J, Hammarén R, Pal C, Östman M, Björlenius B, Flach C-F, Kristiansson E, Fick J, Tysklind M, Larsson DGJ: Elucidating selection processes for antibiotic resistance in sewage treatment plants using metagenomics. Science of the Total Environment, 572, 697–712 (2016). doi: 10.1016/j.scitotenv.2016.06.228 [Paper link]
- Pal C, Bengtsson-Palme J, Kristiansson E, Larsson DGJ: The structure and diversity of human, animal and environmental resistomes. Microbiome, 4, 54 (2016). doi: 10.1186/s40168-016-0199-5 [Paper link]
- Flach C-F, Pal C, Svensson CJ, Kristiansson E, Östman M, Bengtsson-Palme J, Tysklind M, Larsson DGJ: Does antifouling paint select for antibiotic resistance? Science of the Total Environment, 590–591, 461–468 (2017). doi: 10.1016/j.scitotenv.2017.01.213 [Paper link]
- Bengtsson-Palme J, Angelin M, Huss M, Kjellqvist S, Kristiansson E, Palmgren H, Larsson DGJ, Johansson A: The human gut microbiome as a transporter of antibiotic resistance genes between continents. Antimicrobial Agents and Chemotherapy, 59, 10, 6551–6560 (2015). doi: 10.1128/AAC.00933-15 [Paper link]
Published paper: Evaluating taxonomic classification software
Yesterday, Molecular Ecology Resources put online an unedited version of a recent paper which I co-authored. This time, Rodney Richardson at Ohio State University has made a tremendous work of evaluating three taxonomic classification software – the RDP Naïve Bayesian Classifier, RTAX and UTAX – on a set of DNA barcoding regions commonly used for plants, namely the ITS2, and the matK, rbcL, trnL and trnH genes.
In the paper (1), we discuss the results, merits and limitations of the classifiers. In brief, we found that:
- There is a considerable trade-off between accuracy and sensitivity for the classifiers tested, which indicates a need for improved sequence classification tools (2)
- UTAX was superior with respect to error rate, but was exceedingly stringent and thus suffered from a low assignment rate
- The RDP Naïve Bayesian Classifier displayed high sensitivity and low error at the family and order levels, but had a genus-level error rate of 9.6 percent
- RTAX showed high sensitivity at all taxonomic ranks, but at the same time consistently produced the high error rates
- The choice of locus has significant effects on the classification sensitivity of all tested tools
- All classifiers showed strong relationships between database completeness, classification sensitivity and classification accuracy
We believe that the methods of comparison we have used are simple and robust, and thereby provides a methodological and conceptual foundation for future software evaluations. On a personal note, I will thoroughly enjoy working with Rodney and Reed again; I had a great time discussing the ins and outs of taxonomic classification with them! The paper can be found here.
References and notes
- Richardson RT, Bengtsson-Palme J, Johnson RM: Evaluating and Optimizing the Performance of Software Commonly Used for the Taxonomic Classification of DNA Sequence Data. Molecular Ecology Resources, Early view (2016). doi: 10.1111/1755-0998.12628 [Paper link]
- This is something that several classifiers also showed in the evaluation we did for the Metaxa2 paper (3). Interestingly enough, Metaxa2 is better at maintaining high accuracy also as sensitivity is increased.
- Bengtsson-Palme J, Hartmann M, Eriksson KM, Pal C, Thorell K, Larsson DGJ, Nilsson RH: Metaxa2: Improved identification and taxonomic classification of small and large subunit rRNA in metagenomic data. Molecular Ecology Resources, 15, 6, 1403–1414 (2015). doi: 10.1111/1755-0998.12399 [Paper link]