The research leaders of the future are… men
The Swedish Foundation for Strategic Research (SSF) has made public their grants to the research leaders of the future (link in Swedish), aiming to help and promote young researchers with a lot of potential and ambition to build their own research groups within their fields. 18 persons got 10 million SEK each (roughly 1.5 million USD), and also a leadership education. However, SSF obviously believes that men are superior in building and leading research groups, as 14 of the researchers were men (that’s 78%).
It is often argued that the reason that men get more and larger grants than women [1] is that they are more abundant in academia and that the over-representation of men will solve itself given sufficient time. This makes the SSF decisions particularly saddening. These 18 researchers represent the future of Swedish research, and SSF thinks that the research of the future is better of being led by… men. Alarmingly, the foundation’s statements on gender equality (in Swedish) says that (my translation):
The foundation for strategic research views gender equality as something self-evident, that should permeate not only the operations of the foundation, but also all activities that the foundation supports. Thus, the foundation strives towards that all treatment should be gender neutral, and that the under-represented gender should be given priority when other merits are similar. In an equal nation, research resources of men and women should always be taken advantage of, within all areas.
Still, only 20% of the chosen researchers are women. You may think this is a one-time-only event, but no, no, no, it’s much worse than this. In 2005, six of 18 researchers chosen were women (33%), in 2002 six out of 23 (26%), and 2008 six of 20 (30%). It seems that the SSF regards equality to mean 70% men, 30% women. That’s pretty bad for a foundation says it “views gender equality as something self-evident, that should permeate not only the operations of the foundation, but only all activities that the foundation supports.” Obviously, the words on equality are just words, and women still have a long way to go before treated equally by foundations supporting research.
In the long run, this inequality only cements the established norm with men on the top of the research departments. Wennerås and Wold wrote in 2000 that “junior scientists’ frustration at the pace of their scientific productivity is normal at the beginning of their careers, when they do most of the benchwork by themselves. But female scientists tend to remain at this level their entire working lives” [2]. Maybe it would be a good idea for the directors of the SSF to read this, and think about what their actions actually mean for the future of strategic research, and contemplate why women are leaving academia to a much larger extent than men [3]. Because research funders has a huge responsibility for the future of the scientific community.
References
- Wennerås and Wold. Nepotism and sexism in peer-review. Nature (1997) vol. 387 (6631) pp. 341-3
- Wennerås and Wold. A chair of one’s own. Nature (2000) vol. 408 (6813) pp. 647
- Handelsman et al. Careers in science. More women in science. Science (2005) vol. 309 (5738) pp. 1190-1
Raising the bar for genome sequencing
In a recent Nature article (1), Craig Venter and his co-workers at JCVI has not only sequenced one marine bacterium, but 137 different isolates. Their main goal of this study was to better understand the ecology of marine picoplankton in the context of Global Ocean Sampling (GOS) data (2,3). As I see it, there are at least two really interesting things going on here:
First, this is a milestone in sequencing. Were not talking one genome – one article anymore. Were talking one article – 137 new genomes. This vastly raises the bar for any sequencing efforts in the future, but even more importantly, it shifts the focus even further from the actual sequencing to the purpose of the sequencing. One sequenced genome might be interesting enough if it fills a biological knowledge gap, but just sequencing a bacterial strain isn’t worth that much anymore. With the arrival of second- and third-generation sequencing techniques, this development was pretty obvious, but this article is (to my knowledge) the first real proof of that this has finally happened. I expect that five to ten years from now, not sequencing an organism of interest for your research will be viewed as very strange and backwards-looking. “Why didn’t you sequence this?” will be a highly relevant review question for many publications. But also the days when you could write “we here publish for the first time the complete genome sequence of <insert organism name here>” and have that as the central theme for an article will soon be over. Sequencing will simply be reduced to the (valuable) tool it actually is. Which is probably good, as it brings us back to biology again. Articles like this one, where you look at ~200 genomes to investigate ecological questions, are simply providing a more relevant biological perspective than staring at the sequence of one genome in a time when DNA-data is flooding over us.
Second, this is the first (again, to my knowledge) publication where questions arising from metagenomics (2,3,4) has initiated a huge sequencing effort to understand the ecology or the environment to which the metagenome is associated. This highlights a new use of metagenomics as a prospective technique, to mine various environments for interesting features, and then select a few of its inhabitants and look closer at who is responsible for what. With a number of emerging single cell sequencing and visualisation techniques (5,6,7,8) as well as the application of cell sorting approaches to environmental communities (5,9), we can expect metagenomics to play a huge role in organism, strain and protein discovery, but also in determining microbial ecosystem services. Though Venter’s latest article (1) is just a first step towards this new role for metagenomics, it’s a nice example of what (meta)genomics could look like towards the end of this decade, if even not sooner.
- Yooseph et al. Genomic and functional adaptation in surface ocean planktonic prokaryotes. Nature (2010) vol. 468 (7320) pp. 60-6
- Yooseph et al. The Sorcerer II Global Ocean Sampling expedition: expanding the universe of protein families. Plos Biol (2007) vol. 5 (3) pp. e16
- Rusch et al. The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. Plos Biol (2007) vol. 5 (3) pp. e77
- Rusch et al. Characterization of Prochlorococcus clades from iron-depleted oceanic regions. Proceedings of the National Academy of Sciences of the United States of America (2010) pp.
- Woyke et al. Assembling the marine metagenome, one cell at a time. PLoS ONE (2009) vol. 4 (4) pp. e5299
- Woyke et al. One bacterial cell, one complete genome. PLoS ONE (2010) vol. 5 (4) pp. e10314
- Moraru et al. GeneFISH – an in situ technique for linking gene presence and cell identity in environmental microorganisms. Environ Microbiol (2010) pp.
- Lasken. Genomic DNA amplification by the multiple displacement amplification (MDA) method. Biochem Soc Trans (2009) vol. 37 (Pt 2) pp. 450-3
- Mary et al. Metaproteomic and metagenomic analyses of defined oceanic microbial populations using microwave cell fixation and flow cytometric sorting. FEMS microbiology ecology (2010) pp.
Alex Bateman says protein family wiki is coming – but when?
Nature recently had a nice news article on Bio-wikis and biological databases connected to Wikipedia where Alex Bateman says they’re working on a protein-family wiki that will be hosted on Wikipedia, similar to the Rfam wiki, which he talked about at FEBS this summer. I am of course very excited about this, and hope that the new Pfam (?) wiki will come rather sooner than later. As pointed out earlier, the Nature article also underlines the problem of scientific wikis; currently there is no career incentive to get researchers to spend their time editing wiki-articles. That is a shame, and perhaps a system copying the system of Rfam and RNA Biology could help in this direction. The only question is which journal(s) that would be interested in such a commitment to open science…
BLAST Visualization
Perhaps because of my roots in systems biology (or the cause of going there in the first place), I have always had an interest in creating visually appealing images of data, many times in the form of networks. I find that often in bioinformatics, one of the hardest problems is to make information understandable. For example, a BLAST output might say very little about how the genes or proteins are connected to each other, at least to the untrained eye.
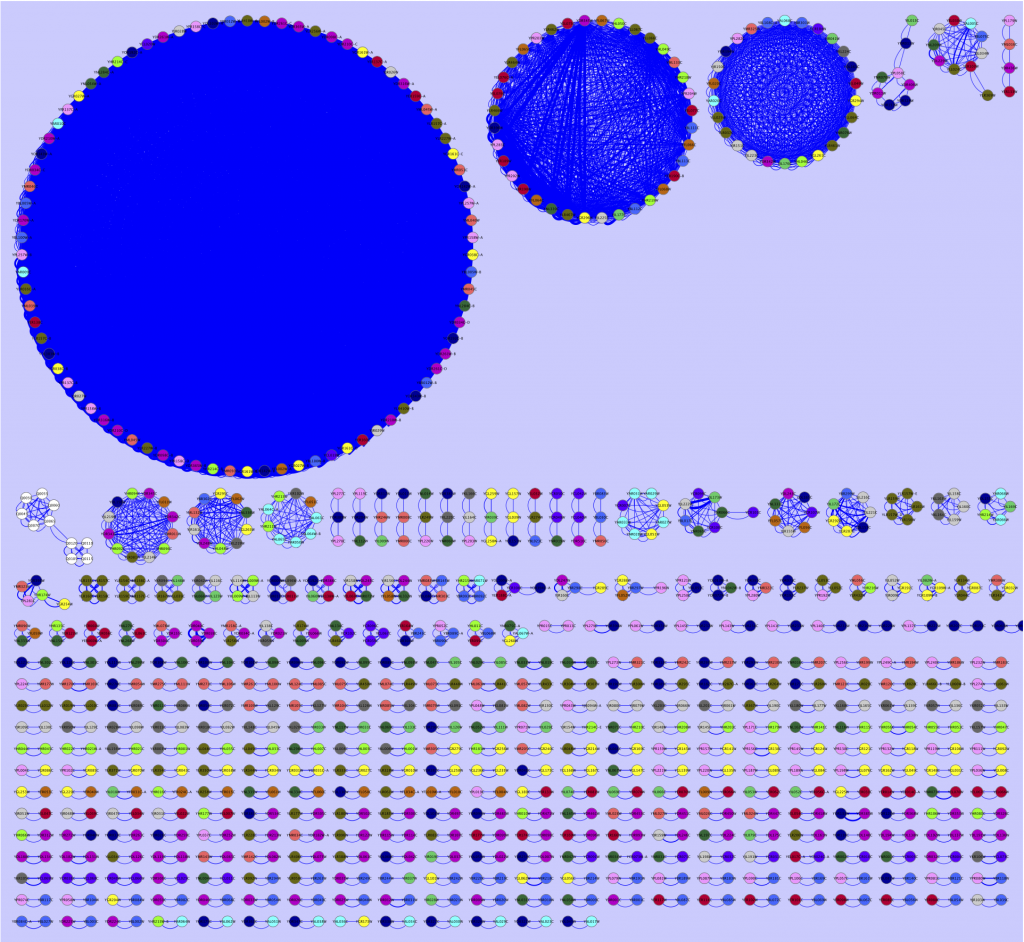
Therefore, during the last weeks I have fiddled around with various ways of viewing interesting portions of BLAST reports. By making all-against-all BLAST searches, and outputting the data in table format (blastall option -m 8), I have been able to extract the hits I am interested in and export them into a Cytoscape compatible format, with some accompanying metadata (scores, e-values, alignment length, etc.). The results are many times pretty unparsable by the eye, rendering them a bit meaningless, but have been more and more interesting as I have put more effort into the extraction script. Just as an example, I here provide a simple map of the best all-against-all matches in the Saccharomyces cerevisiae genome, as a Cytoscape network (click for full size):
The largest circle consists of transposable elements (jumping DNA which inserts itself at multiple locations in the genome, no surprise there is a lot of them, and that these are pretty conserved). The circle to the left of the transposon circle consists of genes located inside the telomeric regions. Why they show such high similarity I do not know, but it seems plausible that the telomere thing could play a role here. The third circle contain mostly members of the seripauperin multigene family, which is also located close to the telomeres. At the bottom you found the gene pairs, that match to each other. You could go on with all the smaller structures as well, but I am no yeast expert, so I will stop here, letting this serve as an example of what a BLAST report really look like.
For this image, I have used a blastn report of all yeast ORFs (taken from yeastgenome.org) as input to my extraction tool, selected Cytoscape compatible output, and used a maximal e-value of 0.00001 and an alignment length of at least 50 nts as criteria to be extracted. I have also pooled the sequences according to chromosome number. The pooling was used to color code the nodes in Cytoscape. The edge width is connected to alignment score, a high score renders a thick line, and a low score causes the line to be thin.
I am still working on the extraction tool and will not provide any code yet. Input would, however, be appreciated. My personal opinion is that in the near future, the overload of newly produced DNA and protein sequences will choke us if do not come up with more intuitive ways of displaying data. I don’t think that the network above is there yet. Still, it conveys information I would not have been able to understand from just looking at the BLAST output. The first attempts to come around the sequence overload problem won’t be the best ones. But we got to start working on visualization methods today, so that we do not end up with sequences over our shoulders in just a few years. Besides, a network image seems much more impressive than a number of lines of text…
A future system for publications?
 I listened to a great talk by Alex Bateman (one of the guys behind Pfam and Rfam, as well as involved in HMMER development) at FEBS yesterday. In addition to talking about the problems of increasing sequence amounts, Alex also provided some reflections on co-operativity and knowledge-sharing – not only among fellow researchers, but also to a wider audience. The starting point of this discussion is Rfam, where the annotation of RNA families is entirely based on a community-driven wiki, tightly integrated with Wikipedia. This means that to make a change in the Rfam annotation, the same change is also made at the corresponding Wikipedia page for this RNA family. And what’s the use of this? Well, as Alex says, for most of the keywords in molecular biology (and I would guess in all of science), the top hit on Google will be a Wikipedia entry. If not, the Wikipedia entry will be in the top ten list of hits, if a good Wiki page exists. This means that Wikipedia is the primary source of scientific information for the general public, as well as many scientists. Wikipedia – not scientific journals.
I listened to a great talk by Alex Bateman (one of the guys behind Pfam and Rfam, as well as involved in HMMER development) at FEBS yesterday. In addition to talking about the problems of increasing sequence amounts, Alex also provided some reflections on co-operativity and knowledge-sharing – not only among fellow researchers, but also to a wider audience. The starting point of this discussion is Rfam, where the annotation of RNA families is entirely based on a community-driven wiki, tightly integrated with Wikipedia. This means that to make a change in the Rfam annotation, the same change is also made at the corresponding Wikipedia page for this RNA family. And what’s the use of this? Well, as Alex says, for most of the keywords in molecular biology (and I would guess in all of science), the top hit on Google will be a Wikipedia entry. If not, the Wikipedia entry will be in the top ten list of hits, if a good Wiki page exists. This means that Wikipedia is the primary source of scientific information for the general public, as well as many scientists. Wikipedia – not scientific journals.
The consequence of this is that to communicate your research subject, you should contribute to its Wikipedia page. In fact, Bateman argues, we have a responsibility as scientists to provide accurate and correct information to the public through the best sources available, which in most cases would be Wikipedia. To put this in perspective (and here I once again borrow Alex’ words), if somebody told you ten years ago that there would be one single internet site that everybody would visit to find scientific information, and where discussion and continuous improvement would be allowed, encouraged and performed, most people would have said that was too good to be true. But that’s what Wikipedia offers. It is time to get rid of the Wiki-sceptisism, and start improving it.
And so, what about the future of publishing? Bateman has worked hard to form an agreement with the journal RNA Biology to integrate the publishing into the process of adding to the easily accessible public information. To have an article on a new RNA family published under the journal’s RNA families track, the family must not only be submitted to the Rfam database, but the authors must also provide a Wikipedia formatted article, which undergo the same peer-review process as the journal article. This ensures high-quality Wikipedia material, as well as making new scientific discoveries public.
I don’t think there’s a long stretch to guess that in the future, more journals and/or funding agencies will take on similar approaches, as researchers and decision-makers discover the importance of correct, publicly available information. The scientific world is slowly moving towards being more open, also for non-scientists. This openness is of extremely high importance in these times of climate scepticism, GMO controversy, extinction of species, and nuclear power debate. For the public to make proper decisions and send a clear message to the politicians, scientists need to be much better at communicating the current state of knowledge, or what many people prefer to call “truth”.
Sex sells – even in biology
Here at FEBS, I am for the first time doing the reflection that sex obviously can sell anything – even biology. With a mixture of disgust and interest of how much more attention it actually brings, I have been watching the two “antibody princess” girls that’s been running around at the conference, trying to sell antibodies. Unfortunately, I have not brought my camera, so all I have is these pretty bad pictures taken with my phone. I will not name the company behind this, as I do not want to function as an inappropriate extra advertising space for them, but it’s interesting to note that the “sex sells” thing has reached into molecular biology. And it makes me wonder what’s next…
Welcome
 My name is Johan Bengtsson-Palme. I am an assistant professor at the Division of Systems Biology at Chalmers University of Technology in Gothenburg and the Sahlgrenska Academy at University of Gothenburg, funded by the Wallenberg DDLS initiative. My research group works with data driven microbiology and microbial ecology, primarily focusing on investigating antibiotic resistance, pathogenesis and interactions in bacterial communities through large-scale experimental work, metagenomics and bioinformatics. I also have an interest in molecular taxonomy and improving the quality of reference databases. You can read more about our research interests here. To contact me, feel free to send an e-mail to my firstname.lastname@microbiology.se
My name is Johan Bengtsson-Palme. I am an assistant professor at the Division of Systems Biology at Chalmers University of Technology in Gothenburg and the Sahlgrenska Academy at University of Gothenburg, funded by the Wallenberg DDLS initiative. My research group works with data driven microbiology and microbial ecology, primarily focusing on investigating antibiotic resistance, pathogenesis and interactions in bacterial communities through large-scale experimental work, metagenomics and bioinformatics. I also have an interest in molecular taxonomy and improving the quality of reference databases. You can read more about our research interests here. To contact me, feel free to send an e-mail to my firstname.lastname@microbiology.se