Published paper: Metaxa
It is a pleasure to annonce that the paper on Metaxa is now available as an Online early article in Antonie van Leeuwenhoek. In short, the paper describes a software tool that is able to extract small subunit (SSU) rRNA sequences from large data sets, such as metagenomes and environmental PCR libraries, and classify them according to bacterial, archaeal, eukaryote, chloroplast or mitochondrial origin. The program makes it easy to distinguish between e.g. the bacterial SSU sequences you like to analyze, and the SSU sequences you would like to remove prior to the analysis (e.g. mitochondrial and chloroplast sequences). This task is particularly important in metagenomics, where sequences can potentially derive from a variety of origins, but bacterial diversity often is the desired target for analysis. The software can be downloaded here, and the article can be read here. I would like to thank all the co-authors on this paper for a brilliant collaboration, and hope to be working with them again.
Reference:
- Bengtsson J, Eriksson KM, Hartmann M, Wang Z, Shenoy BD, Grelet G, Abarenkov K, Petri A, Alm Rosenblad M, Nilsson RH: Metaxa: A software tool for automated detection and discrimination among ribosomal small subunit (12S/16S/18S) sequences of archaea, bacteria, eukaryotes, mitochondria, and chloroplasts in metagenomes and environmental sequencing datasets. Antonie van Leeuwenhoek Journal of Microbiology, 2011, doi:10.1007/s10482-011-9598-6.
From the literature batch…
A random sample of things from this week’s scientific news I think are worth sharing:
Britain is apparently shutting down many of its climate change outreach efforts. I find this very saddening, and see it as an indication of our extreme short-sightedness. We need to put more effort and funding into preserving the environment – not less. In addition, the economic benefits of taking care of the nature around us will probably be much larger than the small sums we save in the short term by not doing anything. We clearly need better incentives to look beyond the next budget and the next election.
The editorial of Nature Reviews Microbiology points the torch on the need for research within basic microbiology, pointing out that “the functions of many genes in the genomes of even the best studied organisms, such as Escherichia coli and Bacillus subtilis, remain unknown. Often these genes do not resemble other, characterized, genes in the databases, allowing for the possibility that interesting new pathways remain to be discovered. (…) if we want to understand how life works at the molecular level, it is crucial to continue and expand basic microbiology research.” I would like to add that a more complete understanding of at least one model organism would drastically increase the accuracy of genome (and metagenome) annotation in new sequencing projects, which today is patchy, to say the least.
Metaxa updated
Just a short note; Metaxa has been updated to version 1.0.1. This incremental version brings two small new features, and a minimal bug fix.
- Added the option to select whether HMMER’s heuristic filtering should be used or not. This can be configured using the –heuristics option:
–heuristics {T or F} : Selects whether to use HMMER’s heuristic filtering, off (F) by default - Removed some redundant information written to the screen, as output to the screen was a bit cluttered.
Bug fix:
- Fixed a rare bug affecting detection sensivity of some SSU sequences.
Of course I would recommend it to every Metaxa user as it fixes a small bug, but the update is not in anyway critical for normal use. The updated version can be downloaded using this link.
Metaxa released
I proudly announce that today Metaxa has been officially released. Metaxa is a a software tool for automated detection and discrimination among ribosomal small subunit (12S/16S/18S) sequences of archaea, bacteria, eukaryotes, mitochondria, and chloroplasts in metagenomes and environmental sequence datasets. We have been working on Metaxa for quite some time, and it has now been in beta for about two months. However, it seems to be stable enough for public consumption. In addition, the software package is today presented in a talk at the SocBiN conference in Helsinki.
A more thorough post on the rationale behind Metaxa, and how it works will follow when I am not occupied by the SocBiN conference. A paper on Metaxa is to be published in the journal Antonie van Leeuwenhoek. The software can be downloaded from here.
Talk, talk, talk…
For those of you who are not already fed up with my writings on biology stuff on the web site, two opportunities to hear me talk in real life has popped up in May. The first is already on May 2nd, on the Open Day in Life Sciences, arranged by the Science Faculty at the University of Gothenburg. I will talk about the search for detoxification systems in metagenomic sequence data (from a collections point of view, as that is the theme for the day). There will also be an opportunity be guided in the herbarium and the botanical garden, plus having lunch and an optional after-work drink at Botaniska Paviljongen. But hurry, last day of admission is tomorrow! Register here.
The second opportunity will be at the SocBiN-2011 bioinformatics conference in Helsinki, on the 12th of May. I will present in the session called “Bioinformatics of Metagenomics”, and talk about a software tool for rRNA classification. I really look forward to this Bioinformatics conference, there are a number of highly prominent and interesting speakers, and I have heard that Helsinki in May is very beautiful. Besides, I am going there with extremely nice people, adding up to potentially being the best biology venue I will attend this spring.
Moving on…
So, last week I started my Ph.D. in Joakim Larsson’s group at the Sahlgrenska Academy. While I am very happy about how things have evolved, I will also miss the ecotox group and the functional genomics group a lot (though both do their research within 10 minutes walking distance from my new place…) I spent last week getting through the usual administrative hassle; getting keys and cards, signing papers, installing bioinformatics software on my new monster of a computer etc. Slowly, the new room starts to feel like it is mine (after nailing phylogenetic trees, my favorite map of the amino acids, and my remember-why-Cytoscape-visualisation-might-not-be-a-good-idea-for-all-network-like-structures poster to the billboard).
So what will this change of positions mean? Will I quit doing research on microbial communities? Of course not! In my new position, my subject of investigation will be bacterial communities subjected to antibiotics. We will look for resistance genes in such communities, and try to answer questions like: How do a high antibiotic selection pressure affect abundance of resistance genes and mobile elements that could facilitate their transfer between bacteria? Can resistance genes found in environmental bacteria be transferred to the microbes of the human gut? Can the environmental bacteria tell us what resistance genes that will be present in clinical situations in the near future? All these questions could, at least partially, be answered by metagenomic approaches and good bioinformatics tools, and my role will be to come up with the solutions provide answers to them.
I am excited over this new project, which involves my favorite subject – metagenomics and community analysis – as well as important factors, such as the clinical connections, the possibility to add pieces to the antibiotic resistance puzzle, and the role of gene and species transfer in resistance development. I also like the fact that I will need to handle high-throughput sequence data, meaning that there will be many opportunities to develop tools, a task I highly enjoy. I think the next couple of years will be an exciting time.
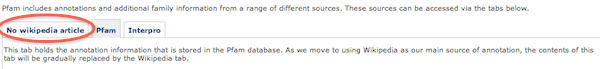
Pfam + Wikipedia – finally!
Browsing the Pfam web site today, I discovered that the database finally has launched its Wikipedia co-ordination efforts.
This has happened along with the 25th release of the Pfam database (released 1st of April), and basically means that Wikipedia articles will be linked to Pfam families. Gradually, this will (hopefully) improve the annotation of Pfam families, which has in many cases been rather poor. The Xfam blog post related to Pfam release 25 says the change will be happening gradually, which might actually be good thing, given the quirks that might pop up.
(…) a major change is that Pfam annotation is now beginning to be co-ordinated via Wikipedia. Unlike Rfam, where every entry has a Wikipedia entry, we expect this to be a more gradual transition for Pfam, so not all entries currently have a corresponding Wikipedia article. For a more detailed discussion, check the help page. We actively encourage the addition of new/updated annotations via Wikipedia as they will appear far quicker than waiting for a Pfam release. If there are articles in Wikipedia that you think correspond to a family, then please mail us!
I have awaited this change for a long time, and is very happy that Pfam has finally taken this step. Congratulations and my sincerest thanks to the Pfam team! Now, let’s go editing!
Software reorganisation
I have reorganised my Software page a little bit, putting the smaller scripts on a separate page, to make the main software page tidier. The content of the pages is the same, and you still find bloutminer and metaorf on the main software page.
BLAST-mining software: bloutminer
I have put some “new” software online. I have had this piece of code lying around for some time but never got to upload it as I didn’t view it as “finished”. It is still not finished, but I would nevertheless like to share it with a wider audience. So, today I introduce bloutminer – the BLAST output mining script I have been using lately. bloutminer allows you to specify e.g. an E-value cutoff, a length cutoff and a percent identity cutoff, and extract a list of the hits satisfying these cutoffs. It takes table output (blastall option -m 8 ) as input. This is the software I used for the BLAST visualisation I have discussed earlier.
I normally use an E-value cutoff of 10 for my BLAST searches, and then extracts hits with bloutminer, allowing me to change the cutoffs at a later stage without redoing the whole BLAST search. You can also “pool” sequences into groups, based on their sequence tags. bloutminer is work in progress, and may contain nasty bugs. It can be found on the Software page. Please improve it at will.
Thesis presentation
I will present my master thesis “Metagenomic Analysis of Marine Periphyton Communities”, on Tuesday the 22nd of March, at 13.00. The presentation will take place in the room Folke Andreasson at Medicinaregatan 11 in Gothenburg. The presentation is open for everyone, but the number of seats are limited.