Published paper: Environmental factors leading to resistance
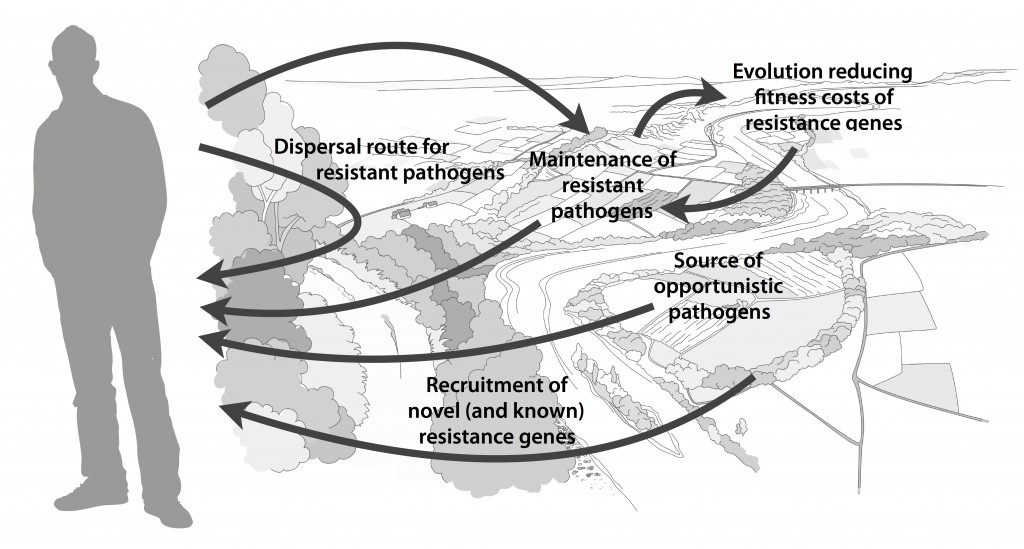
Myself, Joakim Larsson and Erik Kristiansson have written a review on the environmental factors that influence development and spread of antibiotic resistance, which was published today in FEMS Microbiology Reviews. The review (1) builds on thoughts developed in the latter parts of my PhD thesis (2), and seeks to provide a synthesis knowledge gained from different subfields towards the current understanding of evolutionary and ecological processes leading to clinical appearance of resistance genes, as well as the important environmental dispersal barriers preventing spread of resistant pathogens.
We postulate that emergence of novel resistance factors and mobilization of resistance genes are likely to occur continuously in the environment. However, the great majority of such genetic events are unlikely to lead to establishment of novel resistance factors in bacterial populations, unless there is a selection pressure for maintaining them or their fitness costs are negligible. To enable measures to prevent resistance development in the environment, it is therefore critical to investigate under what conditions and to what extent environmental selection for resistance takes place. Selection for resistance is likely less important for the dissemination of resistant bacteria, but will ultimately depend on how well the species or strain in question thrives in the external environment. Metacommunity theory (3,4) suggests that dispersal ability is central to this process, and therefore opportunistic pathogens with their main habitat in the environment may play an important role in the exchange of resistance factors between humans and the environment. Understanding the dispersal barriers hindering this exchange is not only key to evaluate risks, but also to prevent resistant pathogens, as well as novel resistance genes, from reaching humans.
Towards the end of the paper, we suggest certain environments that seem to be more important from a risk management perspective. We also discuss additional problems linked to the development of antibiotic resistance, such as increased evolvability of bacterial genomes (5) and which other types of genes that may be mobilized in the future, should the development continue (1,6). In this review, we also further develop thoughts on the relative risks of re-recruiting and spreading well-known resistance factors already circulating in pathogens, versus recruitment of completely novel resistance genes from environmental bacteria (7). While the latter case is likely to be very rare, and thus almost impossible to quantify the risks for, the consequences of such (potentially one-time) events can be dire.
I personally think that this is one of the best though-through pieces I have ever written, and since it is open access and (in my biased opinion) written in a fairly accessible way, I recommend everyone to read it. It builds on the ecological theories for resistance ecology developed by, among others, Fernando Baquero and José Martinez (8-13). Over the last year, it has been stressed several times at meetings (e.g. at the EDAR conferences in August) that there is a need to develop an ecological framework for antibiotic resistance genes. I think this paper could be one of the foundational pillars on such an endeavor and look forward to see how it will fit into the growing literature on the subject!
References
- Bengtsson-Palme J, Kristiansson E, Larsson DGJ: Environmental factors influencing the development and spread of antibiotic resistance. FEMS Microbiology Reviews, accepted manuscript (2017). doi: 10.1093/femsre/fux053
- Bengtsson-Palme J: Antibiotic resistance in the environment: a contribution from metagenomic studies. Doctoral thesis (medicine), Department of Infectious Diseases, Institute of Biomedicine, Sahlgrenska Academy, University of Gothenburg, 2016. [Link]
- Bengtsson J: Applied (meta)community ecology: diversity and ecosystem services at the intersection of local and regional processes. In: Verhoef HA, Morin PJ (eds.). Community Ecology: Processes, Models, and Applications. Oxford: Oxford University Press, 115–130 (2009).
- Leibold M, Norberg J: Biodiversity in metacommunities: Plankton as complex adaptive systems? Limnology and Oceanography, 1278–1289 (2004).
- Gillings MR, Stokes HW: Are humans increasing bacterial evolvability? Trends in Ecology and Evolution, 27, 346–352 (2012).
- Gillings MR: Evolutionary consequences of antibiotic use for the resistome, mobilome and microbial pangenome. Frontiers in Microbiology, 4, 4 (2013).
- Bengtsson-Palme J, Larsson DGJ: Antibiotic resistance genes in the environment: prioritizing risks. Nature Reviews Microbiology, 13, 369 (2015). doi: 10.1038/nrmicro3399-c1
- Baquero F, Alvarez-Ortega C, Martinez JL: Ecology and evolution of antibiotic resistance. Environmental Microbiology Reports, 1, 469–476 (2009).
- Baquero F, Tedim AP, Coque TM: Antibiotic resistance shaping multi-level population biology of bacteria. Frontiers in Microbiology, 4, 15 (2013).
- Berendonk TU, Manaia CM, Merlin C et al.: Tackling antibiotic resistance: the environmental framework. Nature Reviews Microbiology, 13, 310–317 (2015).
- Hiltunen T, Virta M, Laine A-L: Antibiotic resistance in the wild: an eco-evolutionary perspective. Philosophical Transactions of the Royal Society B: Biological Sciences, 372 (2017) doi: 10.1098/rstb.2016.0039.
- Martinez JL: Bottlenecks in the transferability of antibiotic resistance from natural ecosystems to human bacterial pathogens. Frontiers in Microbiology, 2, 265 (2011).
- Salyers AA, Amábile-Cuevas CF: Why are antibiotic resistance genes so resistant to elimination? Antimicrobial Agents and Chemotherapy, 41, 2321–2325 (1997).
Talk on emission limits in Stockholm
In two weeks time, on the 15th of June, I will participate in a seminar organised by Landstingens nätverk för läkemedel och miljö (the Swedish county council network for pharmaceuticals and environment; the seminar will be held in Swedish) in Stockholm. I will give a talk on our proposed emission limits for antibiotics published last year (the paper is available here), but there will also be talks on wastewater treatment, sustainable pharmaceutical usage and environmental standards for pharmaceuticals. The full program can be found here, and you may register here until June 9. The seminar is free of charge.
And if you are interested in this, I can also recommend the webinar given by Healthcare Without Harm next week (on June 8), which will deal with sustainable procurement as a means to deal with pharmaceutical pollution in the environment. I will at least tune in to hear how the discussion goes here.
Webinar on Antimicrobial Resistance and the Environment
I will give a short talk on our findings related to antibiotic resistance associated with pharmaceutical production facilities in India at a one-hour webinar arranged by Healthcare Without Harm, taking place on Thursday, November 3rd, 10.00 CET. The webinar will discuss “hot-spot” environments in which antimicrobial resistance can emerge, such as areas in which there are poor pharmaceutical manufacturing practices, where expired or unused drugs are disposed of in an inappropriate way (i.e. by flushing them down the toilet or sink, or disposing them in household rubbish), and areas in which pharmaceuticals are used for aquaculture or agriculture. This is an important aspect of the resistance problem, but to date most of the actions taken to tackle the spread of AMR don’t take into account this aspect of antimicrobials released into the environment. The webinar is co-organised by HCWH Europe and HCWH Asia, and aims to raise awareness about the issue of AMR and its environmental impact. It features, apart from myself, Lucas Wiarda (Global Marketing Director & Head of Sustainable Antibiotics Program at DSM Sinochem Pharmaceuticals) and Sister Mercilyn Jabel (Pharmacist at Saint Paul Hospital Cavite, Philippines).
Sign up here to learn about:
- Antibiotic pollution and waste
- Recent findings from India regarding antibiotic discharges in rivers from manufacturers and new mechanisms by which resistance spreads in the environment
- Sustainable antibiotics – how to support the proper and effective use of antibiotics and their responsible production
- How the pharmaceutical industry is addressing the environmental pollution that leads to AMR
- The best practices in managing infectious waste at hospital level
Published opinion piece: Why limit antibiotic pollution?
Me and Joakim Larsson wrote an opinion/summary piece for the APUA Newsletter, issued by the Alliance for Prudent Use of Antibiotics, that was published yesterday (1). The paper is essentially a summary of work included in my PhD thesis, and discusses how to establish minimal selective concentrations of antibiotics for microbial communities (2-4), how to identify risk environments for resistance selection (5-9), and which mitigation strategies that can be implemented (10-12). Partially, we also discussed these issues earlier in our paper in the Medicine Maker (10), but this paper goes deeper into why limiting antibiotic pollution is important to mitigate the accelerating antibiotic resistance problem. I recommend this short summary piece to anyone who would like a brief overview of our research on antibiotic resistance, and think that it can serve as a great starting point for further reading! In addition, this issue of the newsletter features very interesting pieces on reducing antibiotics use (and disposal) outside of the clinics (13) and revival of old antibiotics (14). Please go ahead to the APUA web site and read the entire newsletter!
References
- Bengtsson-Palme J, Larsson DGJ: Why limit antibiotic pollution? The role of environmental selection in antibiotic resistance development. APUA Newsletter, 34, 2, 6-9 (2016). [Paper link].
- Bengtsson-Palme J, Larsson DGJ: Concentrations of antibiotics predicted to select for resistant bacteria: Proposed limits for environmental regulation. Environment International, 86, 140-149 (2016). doi: 10.1016/j.envint.2015.10.015 [Paper link]
- Gullberg E, Cao S, Berg OG, Ilbäck C, Sandegren L, Hughes D, et al.: Selection of resistant bacteria at very low antibiotic concentrations. PLoS Pathogens 7, e1002158 (2011).
- Lundström S, Östman M, Bengtsson-Palme J, Rutgersson C, Thoudal M, Sircar T, Blanck H, Eriksson KM, Tysklind M, Flach C-F, Larsson DGJ: Minimal selective concentrations of tetracycline in complex aquatic bacterial biofilms. Science of the Total Environment, 553, 587–595 (2016). doi: 10.1016/j.scitotenv.2016.02.103
- Bengtsson-Palme J, Boulund F, Fick J, Kristiansson E, Larsson DGJ: Shotgun metagenomics reveals a wide array of antibiotic resistance genes and mobile elements in a polluted lake in India. Frontiers in Microbiology, 5, 648 (2014). doi: 10.3389/fmicb.2014.00648
- Bengtsson-Palme J, Hammarén R, Pal C, Östman M, Björlenius B, Flach C-F, Kristiansson E, Fick J, Tysklind M, Larsson DGJ: Elucidating selection processes for antibiotic resistance in sewage treatment plants using metagenomics. Science of the Total Environment, in press (2016). doi: 10.1016/j.scitotenv.2016.06.228
- Berendonk TU, Manaia CM, Merlin C, Fatta-Kassinos D, Cytryn E, Walsh F, et al.: Tackling antibiotic resistance: the environmental framework. Nature Reviews Microbiology, 13, 310–317 (2015). doi: 10.1038/nrmicro3439
- Martinez JL, Coque TM, Baquero F: What is a resistance gene? Ranking risk in resistomes. Nature Reviews Microbiology 2015, 13:116–123. doi:10.1038/nrmicro3399
- Bengtsson-Palme J, Larsson DGJ: Antibiotic resistance genes in the environment: prioritizing risks. Nature Reviews Microbiology, 13, 369 (2015) doi:10.1038/nrmicro3399‐c1
- Bengtsson-Palme J, Larsson DGJ: Time to limit antibiotic pollution. The Medicine Maker, 0416, 302, 17–18 (2016). [Paper link]
- Ashbolt NJ, Amézquita A, Backhaus T, Borriello P, Brandt KK, Collignon P, et al.: Human Health Risk Assessment (HHRA) for Environmental Development and Transfer of Antibiotic Resistance. Environmental Health Perspectives, 121, 993–1001 (2013)
- Pruden A, Larsson DGJ, Amézquita A, Collignon P, Brandt KK, Graham DW, et al.: Management options for reducing the release of antibiotics and antibiotic resistance genes to the environment. Environmental Health Perspectives, 121, 878–85 (2013).
- Theuretzbacher U: Optimizing the Use of Old Antibiotics — A Global Health Agenda. APUA Newsletter, 34, 2, 10-13 (2016). [Paper link].
- Amábile-Cuevas CF: Antibiotics and Antibiotic Resistance All Around Us. APUA Newsletter, 34, 2, 3-5 (2016). [Paper link].
Environmental pollution with antibiotics leads to resistance
So, on Thursday (May 26th) I will defend my thesis, titled “Antibiotic resistance in the environment: a contribution from metagenomic studies”. I will not dwell into this by writing a novel text, but will instead shamelessly reproduce the press release, which should give a reasonable overview of what I have been doing:
More and more people are infected with antibiotic resistant bacteria. But how do bacteria become resistant? A doctoral thesis from the Centre for Antibiotic Resistance Research at University of Gothenburg has investigated the role of the environment in the development of antibiotic resistance.
“An important question we asked was how low concentrations of antibiotics that can favour the growth of resistant bacteria in the environment”, says Johan Bengtsson-Palme, author of the thesis.
“Based on our analyses, we propose emission limits for 111 antibiotics that should not be exceeded in order to avoid that environmental bacteria become more resistant.”
A starting point to regulate antibiotic pollution
A recent report, commissioned by the British Prime Minister David Cameron, proposes that the emission limits suggested in Johan’s thesis should be used as a starting point to regulate antibiotic pollution from, for example, pharmaceutical production – globally.
“Many people are surprised that such regulations are not already in place, but today it is actually not a crime to discharge wastewater contaminated with large amounts of antibiotics, not even in Europe”, says Johan Bengtsson-Palme.
Resistance genes
In one of the studies in the thesis, the researchers show that resistance genes against a vast range of antibiotics are enriched in an Indian lake polluted by dumping of wastewater from pharmaceutical production.
“It’s scary. Not only do the bacteria carry a multitude of resistance genes. They are also unusually well adapted to share those genes with other bacteria. If a disease-causing bacterium ends up in the lake, it may quickly pick up the genes it needs to become resistant. Since the lake is located close to residential areas, such spread of resistant bacteria to humans is not hard to imagine”, says Johan Bengtsson-Palme.
Spreading by travelers
The thesis also shows that resistant bacteria spread in the intestines of travelers who have visited India or Central Africa, even if the travelers themselves have not become sick.
“That resistant bacteria spread so quickly across the planet highlights that we must adopt a global perspective on the resistance problem”, says Johan Bengtsson-Palme. “Furthermore, it is not enough to reduce the use of antibiotics in healthcare. We must also reduce the use of antibiotics for animals, and try to limit the releases of antibiotics into the environment to try to get control over the growing antibiotic resistance problem before it is too late”.
The thesis Antibiotic resistance in the environment: a contribution from metagenomic studies will be defended on a dissertation on May 26th.
Published paper: Prioritizing antibiotic resistance risks
Late last year, an opinion paper by José Martínez, Teresa Coque and Fernando Baquero was published in Nature Reviews Microbiology (1). In this paper, the authors present a system – resistance readiness conditions (RESCon) – for ranking the risks associated with the detection of antibiotic resistance genes. They also outline the obstacles associated with determining risks presented by antibiotic resistance genes in environmental microbial communities in terms of their potential to transfer to human pathogens. Generally, I am very positive about this paper, which I think is a must-read for anyone who works with antibiotic resistance genes in metagenomes, regardless of it they stem from the human gut or the external environment.
There is, however, one very important aspect that struck me and many other members of our research group as curious: the proposed system assign antibiotic resistance genes already present on mobile genetic elements in human pathogens to the highest risk category (RESCon 1), while resistance genes encoding novel resistance mechanisms not yet been found on mobile elements in a pathogen are considered to be part of lower risk categories. We believe that this system will overestimate the risks associated with well-known resistance factors that are already circulating among human pathogens and under-appreciate the potentially disastrous consequences that the transfer of previously unknown resistance determinants from the environmental resistome could have (exemplified by the rapid clinical spread of the NDM-1 metallo-beta-lactamase gene (2,3)).
With this in mind me and Joakim Larsson wrote a response letter to Nature Reviews Microbiology that went online last monday (4), together with the authors’ reply to us (5). (I strongly suggest that you read the entire original paper (1) before you read the reply (5) to our response letter (4), since Martinez et al. changes the scope slightly from the original paper in their response letter, and these clarifications may (or may not) have been in response to our arguments.)
In our response, we also stress that the abundances of resistance genes, and not only their presence, should be accounted for when estimating risks (although that last point might have been slightly obscured due to the very low word limit). In other words, we think that identifying environmental hotspots for antibiotic resistance genes, where novel resistance genes could be selected for (6,7,8), is of great importance for mitigating public health risks related to environmental antibiotic resistance. Please read our full thoughts on the matter in Nature Reviews Microbiology.
Similar issues will be touched upon in my talk at the EDAR2015 conference later in May. Hope to see you there!
References
- Martinez JL, Coque TM, Baquero F: What is a resistance gene? Ranking risk in resistomes. Nat Rev Microbiol 2015, 13:116–123.
- Kumarasamy KK, et al.: Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis 2010, 10:597–602.
- Walsh TR, Weeks J, Livermore DM, Toleman MA: Dissemination of NDM‐1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect Dis 2011, 11:355–362.
- Bengtsson-Palme J, Larsson DGJ: Antibiotic resistance genes in the environment: prioritizing risks. Nat Rev Microbiol 2015, Advance online publication. doi:10.1038/nrmicro3399‐c1
- Martinez JL, Coque TM, Baquero F: Prioritizing risks of antibiotic resistance genes in all metagenomes. Nat Rev Microbiol 2015, Advance online publication. doi:10.1038/nrmicro3399‐c2
- Kristiansson E, et al.: Pyrosequencing of antibiotic‐contaminated river sediments reveals high levels of resistance and gene transfer elements. PLoS ONE 2011, 6:e17038.
- Bengtsson‐Palme J, Boulund F, Fick J, Kristiansson E, Larsson DGJ: Shotgun metagenomics reveals a wide array of antibiotic resistance genes and mobile elements in a polluted lake in India. Front Microbiol 2014, 5:648.
- Marathe NP, et al.: A treatment plant receiving waste water from multiple bulk drug manufacturers is a reservoir for highly multi‐drug resistant integron‐bearing bacteria. PLoS ONE 2013, 8:e77310.
Indian lake picked up by Indian media
It is nice to see that Indian media has picked up the story about antibiotic resistance genes in the heavily polluted Kazipally lake. In this case, it is the Deccan Chronicle who have been reporting on our findings and briefly interviewed Prof. Joakim Larsson about the study. The issue of pharmaceutical pollution of the environment in drug-producing countries is still rather under-reported and public perception of the problem might be rather low. Therefore, it makes me happy to see an Indian newspaper reporting on the issue. The scientific publication referred to can be found here.
Polluted lake paper in final form
Our paper describing the bacterial community of a polluted lake in India has now been typeset and appears in its final form in Frontiers in Microbiology. If I may say so, I think that the paper turned out to be very goodlooking and it is indeed nice to finally see it in print. The paper describes an unprecedented diversity and abundance of antibiotic resistance genes and genes enabling transfer of DNA between bacteria. We also describe a range of potential novel plasmids from the lake. Finally, the paper briefly describes a new approach to targeted assembly of metagenomic data — TriMetAss — which can be downloaded here.
Reference:
Bengtsson-Palme J, Boulund F, Fick J, Kristiansson E, Larsson DGJ: Shotgun metagenomics reveals a wide array of antibiotic resistance genes and mobile elements in a polluted lake in India. Frontiers in Microbiology, 5, 648 (2014). doi: 10.3389/fmicb.2014.00648
Published paper: Antibiotic resistance genes in a polluted lake
The first work in which I have employed metagenomics to investigate antibiotic resistance has been accepted in Frontiers in Microbiology, and is (at the time of writing) available as a provisional PDF. In the paper (1), which is co-authored by Fredrik Boulund, Jerker Fick, Erik Kristiansson and Joakim Larsson, we have used shotgun metagenomic sequencing of an Indian lake polluted by dumping of waste from pharmaceutical production. We used this data to describe the diversity of antibiotic resistance genes and the genetic context of those, to try to predict their genetic transferability. We found resistance genes against essentially every major class of antibiotics, as well as large abundances of genes responsible for mobilization of genetic material. Resistance genes were estimated to be 7000 times more abundant in the polluted lake than in a Swedish lake included for comparison, where only eight resistance genes were found. The abundances of resistance genes have previously only been matched by river sediment subject to pollution from pharmaceutical production (2). In addition, we describe twenty-six known and twenty-one putative novel plasmids from the Indian lake metagenome, indicating that there is a large potential for horizontal gene transfer through conjugation. Based on the wide range and high abundance of known resistance factors detected, we believe that it is plausible that novel resistance genes are also present in the lake. We conclude that environments polluted with waste from antibiotic manufacturing could be important reservoirs for mobile antibiotic resistance genes. This work further highlights previous findings that pharmaceutical production settings could provide sufficient selection pressure from antibiotics (3) to drive the development of multi-resistant bacteria (4,5), resistance which may ultimately end up in pathogenic species (6,7). The paper can be read in its entirety here.
References:
- Bengtsson-Palme J, Boulund F, Fick J, Kristiansson E, Larsson DGJ: Shotgun metagenomics reveals a wide array of antibiotic resistance genes and mobile elements in a polluted lake in India. Frontiers in Microbiology, Volume 5, Issue 648 (2014). doi: 10.3389/fmicb.2014.00648
- Kristiansson E, Fick J, Janzon A, Grabic R, Rutgersson C, Weijdegård B, Söderström H, Larsson DGJ: Pyrosequencing of antibiotic-contaminated river sediments reveals high levels of resistance and gene transfer elements. PLoS ONE, Volume 6, e17038 (2011). doi:10.1371/journal.pone.0017038.
- Larsson DGJ, de Pedro C, Paxeus N: Effluent from drug manufactures contains extremely high levels of pharmaceuticals. J Hazard Mater, Volume 148, 751–755 (2007). doi:10.1016/j.jhazmat.2007.07.008
- Marathe NP, Regina VR, Walujkar SA, Charan SS, Moore ERB, Larsson DGJ, Shouche YS: A Treatment Plant Receiving Waste Water from Multiple Bulk Drug Manufacturers Is a Reservoir for Highly Multi-Drug Resistant Integron-Bearing Bacteria. PLoS ONE, Volume 8, e77310 (2013). doi:10.1371/journal.pone.0077310
- Johnning A, Moore ERB, Svensson-Stadler L, Shouche YS, Larsson DGJ, Kristiansson E: Acquired genetic mechanisms of a multiresistant bacterium isolated from a treatment plant receiving wastewater from antibiotic production. Appl Environ Microbiol, Volume 79, 7256–7263 (2013). doi:10.1128/AEM.02141-13
- Pruden A, Larsson DGJ, Amézquita A, Collignon P, Brandt KK, Graham DW, Lazorchak JM, Suzuki S, Silley P, Snape JR., et al.: Management options for reducing the release of antibiotics and antibiotic resistance genes to the environment. Environ Health Perspect, Volume 121, 878–885 (2013). doi:10.1289/ehp.1206446
- Finley RL, Collignon P, Larsson DGJ, McEwen SA, Li X-Z, Gaze WH, Reid-Smith R, Timinouni M, Graham DW, Topp E: The scourge of antibiotic resistance: the important role of the environment. Clin Infect Dis, Volume 57, 704–710 (2013). doi:10.1093/cid/cit355