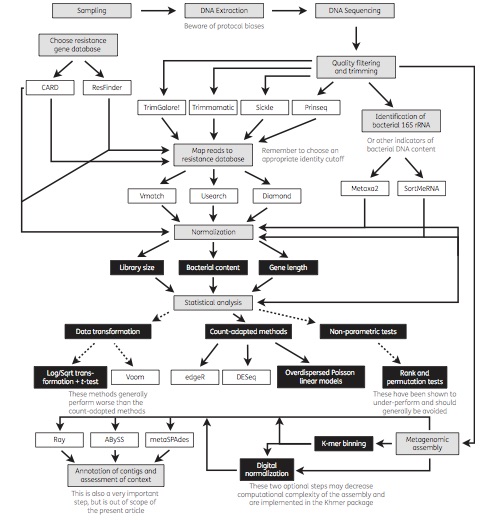
Published paper: Investigating resistomes using metagenomics
Today, a review paper which I wrote together with Joakim Larsson and Erik Kristiansson was published in Journal of Antimicrobial Chemotherapy (1). We have for a long time used metagenomic DNA sequencing to study antibiotic resistance in different environments (2-6), including in the human microbiota (7). Generally, our ultimate purpose has been to assess the risks to human health associated with resistance genes in the environment. However, a multitude of methods exist for metagenomic data analysis, and over the years we have learned that not all methods are suitable for the investigation of resistance genes for this purpose. In our review paper, we describe and discuss current methods for sequence handling, mapping to databases of resistance genes, statistical analysis and metagenomic assembly. We also provide an overview of important considerations related to the analysis of resistance genes, and end by recommending some of the currently used tools, databases and methods that are best equipped to inform research and clinical practice related to antibiotic resistance (see the figure from the paper below). We hope that the paper will be useful to researchers and clinicians interested in using metagenomic sequencing to better understand the resistance genes present in environmental and human-associated microbial communities.
References
- Bengtsson-Palme J, Larsson DGJ, Kristiansson E: Using metagenomics to investigate human and environmental resistomes. Journal of Antimicrobial Chemotherapy, advance access (2017). doi: 10.1093/jac/dkx199 [Paper link]
- Bengtsson-Palme J, Boulund F, Fick J, Kristiansson E, Larsson DGJ: Shotgun metagenomics reveals a wide array of antibiotic resistance genes and mobile elements in a polluted lake in India. Frontiers in Microbiology, 5, 648 (2014). doi: 10.3389/fmicb.2014.00648 [Paper link]
- Lundström S, Östman M, Bengtsson-Palme J, Rutgersson C, Thoudal M, Sircar T, Blanck H, Eriksson KM, Tysklind M, Flach C-F, Larsson DGJ: Minimal selective concentrations of tetracycline in complex aquatic bacterial biofilms. Science of the Total Environment, 553, 587–595 (2016). doi: 10.1016/j.scitotenv.2016.02.103 [Paper link]
- Bengtsson-Palme J, Hammarén R, Pal C, Östman M, Björlenius B, Flach C-F, Kristiansson E, Fick J, Tysklind M, Larsson DGJ: Elucidating selection processes for antibiotic resistance in sewage treatment plants using metagenomics. Science of the Total Environment, 572, 697–712 (2016). doi: 10.1016/j.scitotenv.2016.06.228 [Paper link]
- Pal C, Bengtsson-Palme J, Kristiansson E, Larsson DGJ: The structure and diversity of human, animal and environmental resistomes. Microbiome, 4, 54 (2016). doi: 10.1186/s40168-016-0199-5 [Paper link]
- Flach C-F, Pal C, Svensson CJ, Kristiansson E, Östman M, Bengtsson-Palme J, Tysklind M, Larsson DGJ: Does antifouling paint select for antibiotic resistance? Science of the Total Environment, 590–591, 461–468 (2017). doi: 10.1016/j.scitotenv.2017.01.213 [Paper link]
- Bengtsson-Palme J, Angelin M, Huss M, Kjellqvist S, Kristiansson E, Palmgren H, Larsson DGJ, Johansson A: The human gut microbiome as a transporter of antibiotic resistance genes between continents. Antimicrobial Agents and Chemotherapy, 59, 10, 6551–6560 (2015). doi: 10.1128/AAC.00933-15 [Paper link]
Published paper: The global resistome
Late yesterday, Microbiome put online our most recent work, covering the resistomes to antibiotics, biocides and metals across a vast range of environments. In the paper (1), we perform the largest characterization of resistance genes, mobile genetic elements (MGEs) and bacterial taxonomic compositions to date, covering 864 different metagenomes from humans (350), animals (145) and external environments such as soil, water, sewage, and air (369 in total). All the investigated metagenomes were sequenced to at least 10 million reads each, using Illumina technology, making the results more comparable across environments than in previous studies (2-4).
We found that the environment types had clear differences both in terms of resistance profiles and bacterial community composition. Humans and animals hosted microbial communities with limited taxonomic diversity as well as low abundance and diversity of biocide/metal resistance genes and MGEs. On the contrary, the abundance of ARGs was relatively high in humans and animals. External environments, on the other hand, showed high taxonomic diversity and high diversity of biocide/metal resistance genes and MGEs. Water, sediment and soil generally carried low relative abundance and few varieties of known ARGs, whereas wastewater and sludge were on par with the human gut. The environments with the largest relative abundance and diversity of ARGs, including genes encoding resistance to last resort antibiotics, were those subjected to industrial antibiotic pollution and air samples from a Beijing smog event.
A paper investigating this vast amount of data is of course hard to describe in a blog post, so I strongly suggest the interested reader to head over to Microbiome’s page and read the full paper (1). However, here’s a ver short summary of the findings:
- The median relative abundance of ARGs across all environments was 0.035 copies per bacterial 16S rRNA
- Antibiotic-polluted environments have (by far) the highest abundances of ARGs
- Urban air samples carried high abundance and diversity of ARGs
- Human microbiota has high abundance and diversity of known ARGs, but low taxonomic diversity compared to the external environment
- The human and animal resistomes are dominated by tetracycline resistance genes
- Over half of the ARGs were only detected in external environments, while 20.5 % were found in human, animal and at least one of the external environments
- Biocide and metal resistance genes are more common in external environments than in the human microbiota
- Human microbiota carries low abundance and richness of MGEs compared to most external environments
Importantly, less than 1.5 % of all detected ARGs were found exclusively in the human microbiome. At the same time, 57.5 % of the known ARGs were only detected in metagenomes from environmental samples, despite that the majority of the investigated ARGs were initially encountered in pathogens. Still, our analysis suggests that most of these genes are relatively rare in the human microbiota. Environmental samples generally contained a wider distribution of resistance genes to a more diverse set of antibiotics classes. For example, the relative abundance of beta-lactam resistance genes was much larger in external environments than in human and animal microbiomes. This suggests that the external environment harbours many more varieties of resistance genes than the ones currently known from the clinic. Indeed, functional metagenomics has resulted in the discovery of many novel ARGs in external environments (e.g. 5). This all fits well with an overall much higher taxonomic diversity of environmental microbial communities. In terms of consequences associated with the potential transfer of ARGs to human pathogens, we argue that unknown resistance genes are of greater concern than those already known to circulate among human-associated bacteria (6).
This study describes the potential for many external environments, including those subjected to pharmaceutical pollution, air and wastewater/sludge, to serve as hotspots for resistance development and/or transmission of ARGs. In addition, our results indicate that these environments may play important roles in the mobilization of yet unknown ARGs and their further transmission to human pathogens. To provide guidance for risk-reducing actions we – based on this study – suggest strict regulatory measures of waste discharges from pharmaceutical industries and encourage more attention to air in the transmission of antibiotic resistance (1).
References
- Pal C, Bengtsson-Palme J, Kristiansson E, Larsson DGJ: The structure and diversity of human, animal and environmental resistomes. Microbiome, 4, 54 (2016). doi: 10.1186/s40168-016-0199-5
- Durso LM, Miller DN, Wienhold BJ. Distribution and quantification of antibiotic resistant genes and bacteria across agricultural and non-agricultural metagenomes. PLoS One. 2012;7:e48325.
- Nesme J, Delmont TO, Monier J, Vogel TM. Large-scale metagenomic-based study of antibiotic resistance in the environment. Curr Biol. 2014;24:1096–100.
- Fitzpatrick D, Walsh F. Antibiotic resistance genes across a wide variety of metagenomes. FEMS Microbiol Ecol. 2016. doi:10.1093/femsec/fiv168.
- Allen HK, Moe LA, Rodbumrer J, Gaarder A, Handelsman J. Functional metagenomics reveals diverse β-lactamases in a remote Alaskan soil. ISME J. 2009;3:243–51.
- Bengtsson-Palme J, Larsson DGJ: Antibiotic resistance genes in the environment: prioritizing risks. Nature Reviews Microbiology, 13, 369 (2015). doi: 10.1038/nrmicro3399-c1
Published paper: Antibiotic resistance in sewage treatment plants
 After a long wait (1), Science of the Total Environment has finally decided to make our paper on selection of antibiotic resistance genes in sewage treatment plants (STPs) available (2). STPs are often suggested to be “hotspots” for emergence and dissemination of antibiotic-resistant bacteria (3-6). However, we actually do not know if the selection pressures within STPs, that can be caused either by residual antibiotics or other co-selective agents, are sufficiently large to specifically promote resistance. To better understand this, we used shotgun metagenomic sequencing of samples from different steps of the treatment process (incoming water, treated water, primary sludge, recirculated sludge and digested sludge) in three Swedish STPs in the Stockholm area to characterize the frequencies of resistance genes to antibiotics, biocides and metal, as well as mobile genetic elements and taxonomic composition. In parallel, we also measured concentrations of antibiotics, biocides and metals.
After a long wait (1), Science of the Total Environment has finally decided to make our paper on selection of antibiotic resistance genes in sewage treatment plants (STPs) available (2). STPs are often suggested to be “hotspots” for emergence and dissemination of antibiotic-resistant bacteria (3-6). However, we actually do not know if the selection pressures within STPs, that can be caused either by residual antibiotics or other co-selective agents, are sufficiently large to specifically promote resistance. To better understand this, we used shotgun metagenomic sequencing of samples from different steps of the treatment process (incoming water, treated water, primary sludge, recirculated sludge and digested sludge) in three Swedish STPs in the Stockholm area to characterize the frequencies of resistance genes to antibiotics, biocides and metal, as well as mobile genetic elements and taxonomic composition. In parallel, we also measured concentrations of antibiotics, biocides and metals.
We found that only the concentrations of tetracycline and ciprofloxacin in the influent water were above those that we predict to cause resistance selection (7). However, there was no consistent enrichment of resistance genes to any particular class of antibiotics in the STPs, neither for biocide and metal resistance genes. Instead, the most substantial change of the bacterial communities compared to human feces (sampled from Swedes in another study of ours (8)) occurred already in the sewage pipes, and was manifested by a strong shift from obligate to facultative anaerobes. Through the treatment process, resistance genes against antibiotics, biocides and metals were not reduced to the same extent as fecal bacteria were.
Worryingly, the OXA-48 beta-lactamase gene was consistently enriched in surplus and digested sludge. OXA-48 is still rare in Swedish clinical isolates (9), but provides resistance to carbapenems, one of our most critically important classes of antibiotics. However, taken together metagenomic sequencing did not provide clear support for any specific selection of antibiotic resistance. Rather, since stronger selective forces affect gross taxonomic composition, and thereby also resistance gene abundances, it is very hard to interpret the metagenomic data from a risk-for-selection perspective. We therefore think that comprehensive analyses of resistant vs. non-resistant strains within relevant species are warranted.
Taken together, the main take-home messages of the paper (2) are:
- There were no apparent evidence for direct selection of resistance genes by antibiotics or co-selection by biocides or metals
- Abiotic factors (mostly oxygen availability) strongly shape taxonomy and seems to be driving changes of resistance genes
- Metagenomic and/or PCR-based community studies may not be sufficiently sensitive to detect selection effects, as important shifts towards resistant may occur within species and not on the community level
- The concentrations of antibiotics, biocides and metals were overall reduced, but not removed in STPs. Incoming concentrations of antibiotics in Swedish STPs are generally low
- Resistance genes are overall reduced through the treatment process, but far from eliminated
References and notes
- Okay, those who takes notes know that I have already complained once before on Science of the Total Environment’s ridiculously long production handling times. But, seriously, how can a journal’s production team return the proofs for after three days of acceptance, and then wait seven weeks before putting the final proofs online? I still wonder what is going on beyond the scenes, which is totally obscure because the production office also refuses to respond to e-mails. Not a nice publishing experience this time either.
- Bengtsson-Palme J, Hammarén R, Pal C, Östman M, Björlenius B, Flach C-F, Kristiansson E, Fick J, Tysklind M, Larsson DGJ: Elucidating selection processes for antibiotic resistance in sewage treatment plants using metagenomics. Science of the Total Environment, in press (2016). doi: 10.1016/j.scitotenv.2016.06.228 [Paper link]
- Rizzo L, Manaia C, Merlin C, Schwartz T, Dagot C, Ploy MC, Michael I, Fatta-Kassinos D: Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: a review. Science of the Total Environment, 447, 345–360 (2013). doi: 10.1016/j.scitotenv.2013.01.032
- Laht M, Karkman A, Voolaid V, Ritz C, Tenson T, Virta M, Kisand V: Abundances of Tetracycline, Sulphonamide and Beta-Lactam Antibiotic Resistance Genes in Conventional Wastewater Treatment Plants (WWTPs) with Different Waste Load. PLoS ONE, 9, e103705 (2014). doi: 10.1371/journal.pone.0103705
- Yang Y, Li B, Zou S, Fang HHP, Zhang T: Fate of antibiotic resistance genes in sewage treatment plant revealed by metagenomic approach. Water Research, 62, 97–106 (2014). doi: 10.1016/j.watres.2014.05.019
- Berendonk TU, Manaia CM, Merlin C, Fatta-Kassinos D, Cytryn E, Walsh F, et al.: Tackling antibiotic resistance: the environmental framework. Nature Reviews Microbiology, 13, 310–317 (2015). doi: 10.1038/nrmicro3439
- Bengtsson-Palme J, Larsson DGJ: Concentrations of antibiotics predicted to select for resistant bacteria: Proposed limits for environmental regulation. Environment International, 86, 140–149 (2016). doi: 10.1016/j.envint.2015.10.015
- Bengtsson-Palme J, Angelin M, Huss M, Kjellqvist S, Kristiansson E, Palmgren H, Larsson DGJ, Johansson A: The human gut microbiome as a transporter of antibiotic resistance genes between continents. Antimicrobial Agents and Chemotherapy, 59, 10, 6551–6560 (2015). doi: 10.1128/AAC.00933-15
- Hellman J, Aspevall O, Bengtsson B, Pringle M: SWEDRES-SVARM 2014. Consumption of antimicrobials and occurrence of antimicrobial resistance in Sweden. Public Health Agency of Sweden and National Veterinary Institute, Solna/Uppsala, Sweden. Report No.: 14027. Available from: http://www.folkhalsomyndigheten.se/publicerat-material/ (2014)
Published paper: Co-occurences of resistance genes across bacteria
Yesterday, a paper I co-authored with my colleagues Chandan Pal, Erik Kristiansson and Joakim Larsson on the co-occurences of resistance genes against antibiotics, biocides and metals in bacterial genomes and plasmids became published in BMC Genomics. In this paper (1) we utilize the publicly available, fully sequenced, genomes and plasmids in GenBank to investigate the co-occurence network of resistance genes, to better understand risks for co-selection for resistance against different types of compounds. In short, the findings of the paper are that:
- ARGs are associated with BMRG-carrying bacteria and the co-selection potential of biocides and metals is specific towards certain antibiotics
- Clinically important genera host the largest numbers of ARGs and BMRGs and those also have the highest co-selection potential
- Bacteria isolated from human and domestic animal origins have the highest co-selection potential
- Plasmids with co-selection potential tend to be conjugative and carry toxin-antitoxin systems
- Mercury and QACs are potential co-selectors of ARGs on plasmids, however BMRGs are common on chromosomes and could still have indirect co-selection potential
- 14 percent of bacteria and more than 70% of the plasmids completely lacked resistance genes
This analysis was possible thanks to the BacMet database of antibacterial biocide and metal resistance genes, published about two years ago (2). The visualization of the plasmid co-occurence network we ended up with can be seen below. Note the strong connection between the mercury resistance mer operon and the antibiotic resistance genes to the right.
On a side note, it is interesting to note that the underrepresentation of detoxification systems in marine environments we noted last year (3) still seems to hold for genomes (and particularly plasmids), supporting the genome streamlining hypothesis (4).
References:
- Pal C, Bengtsson-Palme J, Kristiansson E, Larsson DGJ: Co-occurrence of resistance genes to antibiotics, biocides and metals reveals novel insights into their co-selection potential. BMC Genomics, 16, 964 (2015). doi: 10.1186/s12864-015-2153-5 [Paper link]
- Pal C, Bengtsson-Palme J, Rensing C, Kristiansson E, Larsson DGJ: BacMet: Antibacterial Biocide and Metal Resistance Genes Database. Nucleic Acids Research, 42, D1, D737-D743 (2014). doi: 10.1093/nar/gkt1252 [Paper link]
- Bengtsson-Palme J, Alm Rosenblad M, Molin M, Blomberg A: Metagenomics reveals that detoxification systems are underrepresented in marine bacterial communities. BMC Genomics, 15, 749 (2014). doi: 10.1186/1471-2164-15-749 [Paper link]
- Giovannoni SJ, Cameron TJ, Temperton B: Implications of streamlining theory for microbial ecology. ISME Journal, 8, 1553-1565 (2014).
A good-looking version of the Travel and Resistance paper
The paper we published in August on travelers carrying resistance genes with them in their gut microbiota has now been typeset and got proper volume and issue numbers assigned to it in Antimicrobial Agents and Chemotherapy. Take a look at it, I personally think it’s quite good-looking.
Also, if you understand Swedish, here is an interview with me broadcasted on Swedish Radio last month about this study and the consequences of it.
The new citation for the paper is:
- Bengtsson-Palme J, Angelin M, Huss M, Kjellqvist S, Kristiansson E, Palmgren H, Larsson DGJ, Johansson A: The human gut microbiome as a transporter of antibiotic resistance genes between continents. Antimicrobial Agents and Chemotherapy, 59, 10, 6551-6560 (2015). doi: 10.1128/AAC.00933-15 [Paper link]
Published paper: Travel spreads resistance genes
Earlier today, my most recent paper (1) became available online, describing resistance gene patterns in the gut microbiota of Swedes before and after travel to the Indian peninsula and central Africa. In this work, we have used metagenomic sequencing of the intestinal microbiome of Swedish students returning from exchange programs to show that the abundance of antibiotic resistance genes in several classes are increased after travel. This work reiterates the findings of several papers describing uptake of resistant bacteria (2-8) or resistance genes (9-11) after travel to destinations with worse resistance situation.
Our paper is important because it:
- Addresses the abundance of a vast range of resistance genes (more than 300).
- Finds evidence for that the overall relative abundance of antibiotic resistance genes increased after travel, without any intake of antibiotics.
- Shows that the sensitivity of metagenomics was, despite very deep sequencing efforts, not sufficient to detect acquisition of the low-abundant (CTX-M) resistance genes responsible for observed ESBL phenotypes.
- Reveals a “core resistome” of resistance genes that are more or less omnipresent, and remain relatively stable regardless of travel, while changes seem to occur in the more variable part of the resistome.
- Hints at increased abundance of Proteobacteria after travel, although this increase could not specifically be linked to resistance gene increases.
- Uses de novo metagenomic assembly to physically link resistance genes in the same sample, giving hints of co-resistance patterns in the gut microbiome.
The paper was a collaboration with Martin Angelin, Helena Palmgren and Anders Johansson at Umeå University, and was made possible by bioinformatics support from SciLifeLab in Stockholm. I highly recommend reading it as a complement to e.g. the Forslund et al. paper (12) describing country-specific antibiotic resistance patterns in the gut microbiota.
Taken together, this study offers a broadened perspective on how the antibiotic resistance potential of the human gut microbiome changes after travel, providing an independent complement to previous studies targeting a limited number of bacterial species or antibiotic resistance genes. Understanding how resistance genes travels the globe is hugely important, since resistance in principle only need to appear in a pathogen once; improper hygiene and travel may then spread novel resistance genes across continents rapidly (13,14).
References
- Bengtsson-Palme J, Angelin M, Huss M, Kjellqvist S, Kristiansson E, Palmgren H, Larsson DGJ, Johansson A: The human gut microbiome as a transporter of antibiotic resistance genes between continents. Antimicrob Agents Chemother Accepted manuscript posted online (2015). doi: 10.1128/AAC.00933-15 [Paper link]
- Gaarslev K, Stenderup J: Changes during travel in the composition and antibiotic resistance pattern of the intestinal Enterobacteriaceae flora: results from a study of mecillinam prophylaxis against travellers’ diarrhoea. Curr Med Res Opin 9:384–387 (1985).
- Paltansing S, Vlot JA, Kraakman MEM, Mesman R, Bruijning ML, Bernards AT, Visser LG, Veldkamp KE: Extended-spectrum β-lactamase-producing enterobacteriaceae among travelers from the Netherlands. Emerging Infect. Dis. 19:1206–1213 (2013).
- Ruppé E, Armand-Lefèvre L, Estellat C, El-Mniai A, Boussadia Y, Consigny PH, Girard PM, Vittecoq D, Bouchaud O, Pialoux G, Esposito-Farèse M, Coignard B, Lucet JC, Andremont A, Matheron S: Acquisition of carbapenemase-producing Enterobacteriaceae by healthy travellers to India, France, February 2012 to March 2013. Euro Surveill. 19 (2014).
- Kennedy K, Collignon P: Colonisation with Escherichia coli resistant to “critically important” antibiotics: a high risk for international travellers. Eur J Clin Microbiol Infect Dis 29:1501–1506 (2010).
- Tham J, Odenholt I, Walder M, Brolund A, Ahl J, Melander E: Extended-spectrum beta-lactamase-producing Escherichia coli in patients with travellers’ diarrhoea. Scand. J. Infect. Dis. 42:275–280 (2010).
- Östholm-Balkhed Å, Tärnberg M, Nilsson M, Nilsson LE, Hanberger H, Hällgren A, Travel Study Group of Southeast Sweden: Travel-associated faecal colonization with ESBL-producing Enterobacteriaceae: incidence and risk factors. J Antimicrob Chemother 68:2144–2153 (2013).
- Kantele A, Lääveri T, Mero S, Vilkman K, Pakkanen SH, Ollgren J, Antikainen J, Kirveskari J: Antimicrobials increase travelers’ risk of colonization by extended-spectrum betalactamase-producing enterobacteriaceae. Clin Infect Dis 60:837–846 (2015).
- von Wintersdorff CJH, Penders J, Stobberingh EE, Oude Lashof AML, Hoebe CJPA, Savelkoul PHM, Wolffs PFG: High rates of antimicrobial drug resistance gene acquisition after international travel, The Netherlands. Emerging Infect. Dis. 20:649–657 (2014).
- Tängdén T, Cars O, Melhus A, Löwdin E: Foreign travel is a major risk factor for colonization with Escherichia coli producing CTX-M-type extended-spectrum beta-lactamases: a prospective study with Swedish volunteers. Antimicrob Agents Chemother 54:3564–3568 (2010).
- Dhanji H, Patel R, Wall R, Doumith M, Patel B, Hope R, Livermore DM, Woodford N: Variation in the genetic environments of bla(CTX-M-15) in Escherichia coli from the faeces of travellers returning to the United Kingdom. J Antimicrob Chemother 66:1005–1012 (2011).
- Forslund K, Sunagawa S, Kultima JR, Mende DR, Arumugam M, Typas A, Bork P: Country-specific antibiotic use practices impact the human gut resistome. Genome Res 23:1163–1169 (2013).
- Bengtsson-Palme J, Larsson DGJ: Antibiotic resistance genes in the environment: prioritizing risks. Nat Rev Microbiol 13:396 (2015).
- Larsson DGJ: Antibiotics in the environment. Ups J Med Sci 119:108–112 (2014).
Looking for a super-interesting job in bioinformatics?
If you’re looking for super-interesting jobs within bioinformatics, you don’t need to look any further. Instead, you should apply for a position at 1928 Diagnostics here in Gothenburg and join them in the fight against antibiotic resistant bacteria. The position is in the development team and the deadline for application is December 19. All the details can be found here.
Published paper: Antibiotic resistance genes in a polluted lake
The first work in which I have employed metagenomics to investigate antibiotic resistance has been accepted in Frontiers in Microbiology, and is (at the time of writing) available as a provisional PDF. In the paper (1), which is co-authored by Fredrik Boulund, Jerker Fick, Erik Kristiansson and Joakim Larsson, we have used shotgun metagenomic sequencing of an Indian lake polluted by dumping of waste from pharmaceutical production. We used this data to describe the diversity of antibiotic resistance genes and the genetic context of those, to try to predict their genetic transferability. We found resistance genes against essentially every major class of antibiotics, as well as large abundances of genes responsible for mobilization of genetic material. Resistance genes were estimated to be 7000 times more abundant in the polluted lake than in a Swedish lake included for comparison, where only eight resistance genes were found. The abundances of resistance genes have previously only been matched by river sediment subject to pollution from pharmaceutical production (2). In addition, we describe twenty-six known and twenty-one putative novel plasmids from the Indian lake metagenome, indicating that there is a large potential for horizontal gene transfer through conjugation. Based on the wide range and high abundance of known resistance factors detected, we believe that it is plausible that novel resistance genes are also present in the lake. We conclude that environments polluted with waste from antibiotic manufacturing could be important reservoirs for mobile antibiotic resistance genes. This work further highlights previous findings that pharmaceutical production settings could provide sufficient selection pressure from antibiotics (3) to drive the development of multi-resistant bacteria (4,5), resistance which may ultimately end up in pathogenic species (6,7). The paper can be read in its entirety here.
References:
- Bengtsson-Palme J, Boulund F, Fick J, Kristiansson E, Larsson DGJ: Shotgun metagenomics reveals a wide array of antibiotic resistance genes and mobile elements in a polluted lake in India. Frontiers in Microbiology, Volume 5, Issue 648 (2014). doi: 10.3389/fmicb.2014.00648
- Kristiansson E, Fick J, Janzon A, Grabic R, Rutgersson C, Weijdegård B, Söderström H, Larsson DGJ: Pyrosequencing of antibiotic-contaminated river sediments reveals high levels of resistance and gene transfer elements. PLoS ONE, Volume 6, e17038 (2011). doi:10.1371/journal.pone.0017038.
- Larsson DGJ, de Pedro C, Paxeus N: Effluent from drug manufactures contains extremely high levels of pharmaceuticals. J Hazard Mater, Volume 148, 751–755 (2007). doi:10.1016/j.jhazmat.2007.07.008
- Marathe NP, Regina VR, Walujkar SA, Charan SS, Moore ERB, Larsson DGJ, Shouche YS: A Treatment Plant Receiving Waste Water from Multiple Bulk Drug Manufacturers Is a Reservoir for Highly Multi-Drug Resistant Integron-Bearing Bacteria. PLoS ONE, Volume 8, e77310 (2013). doi:10.1371/journal.pone.0077310
- Johnning A, Moore ERB, Svensson-Stadler L, Shouche YS, Larsson DGJ, Kristiansson E: Acquired genetic mechanisms of a multiresistant bacterium isolated from a treatment plant receiving wastewater from antibiotic production. Appl Environ Microbiol, Volume 79, 7256–7263 (2013). doi:10.1128/AEM.02141-13
- Pruden A, Larsson DGJ, Amézquita A, Collignon P, Brandt KK, Graham DW, Lazorchak JM, Suzuki S, Silley P, Snape JR., et al.: Management options for reducing the release of antibiotics and antibiotic resistance genes to the environment. Environ Health Perspect, Volume 121, 878–885 (2013). doi:10.1289/ehp.1206446
- Finley RL, Collignon P, Larsson DGJ, McEwen SA, Li X-Z, Gaze WH, Reid-Smith R, Timinouni M, Graham DW, Topp E: The scourge of antibiotic resistance: the important role of the environment. Clin Infect Dis, Volume 57, 704–710 (2013). doi:10.1093/cid/cit355
PhD position: Come and work with us!
If you are thinking about doing a PhD and think that bioinformatics and antibiotic resistance is a cool subject, then now is your chance to come and join us for the next four years! There is a PhD position open i Joakim Larsson’s group, which means that if you get the job you will work with me, Joakim Larsson, Erik Kristiansson, Ørjan Samuelsen and Carl-Fredrik Flach on a super-interesting project relating to discovery of novel beta-lactamase genes (NoCURE). The project aims to better understand where, how and under what circumstances these genetic transfer events take place, in order to provide opportunities to limit or delay resistance development and thus increase the functional lifespan of precious antibiotics. The lion’s share of the work will be related to interpreting large-scale sequencing data generated by collaborators within the project; both genome sequencing and metagenomic data.
This is a great opportunity to prove your bioinformatics skills and use them for something urgently important. Full details about the position can be found here.
PhD position with Erik Kristiansson
If you’re looking for a PhD position in bioinformatics, working with antibiotic resistance, there’s an opening in Erik Krisiansson’s (best bioinformatician in Gothenburg? I think so) group. To apply you need to have a master’s level degree in bioinformatics, mathematical statistics, mathematics, computer science, physics, molecular biology or any equivalent topic, obtained latest June 2014. If you’re a master student and want to join us, this is your chance! You can read more and apply for the position here.