My ISME talk on EMBARK
Ákos Kovács had the brilliant idea of putting up a temporary resource for things you bring up in a talk that you can point people to. I did not do this before my talk at ISME today, but I thought the idea was so good, so here’s a summary and collection of my ISME short-talk on the EMBARK outcomes today:
- More information on EMBARK and its successor SEARCHER can be found on the project website, here: http://antimicrobialresistance.eu Importantly, this is a team effort over four years and I only touched on a few selected things
- Within the project we have looked at typical background levels of antibiotic resistance in the environment. We have already published some of these results (for qPCR abundances) in Abramova et al. 2023
- The average resistance gene in the average environment is present in ~1 in 1000 bacteria, but the variation between different genes is huge
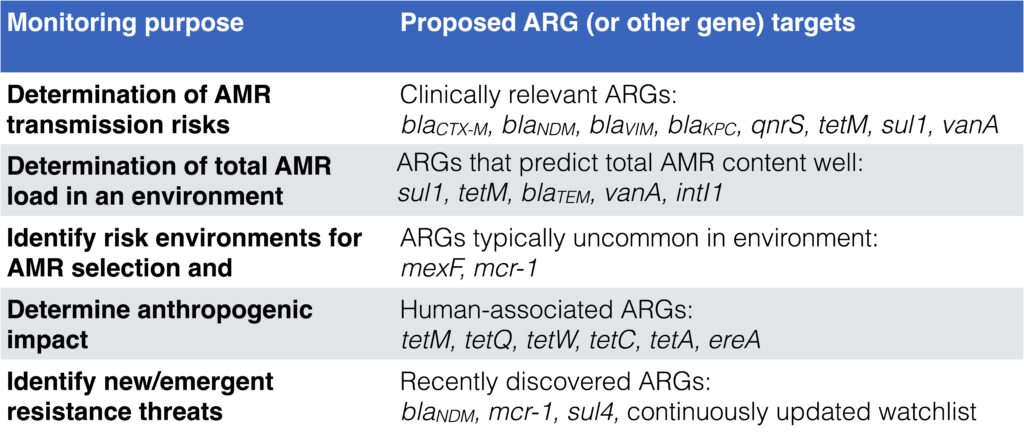
- Depending on monitoring goal, different target genes are relevant to use. See this table adapted from Abramova et al. 2023:
- We have also tried to make different monitoring methods for environmental AMR comparable. Those mentioned in the talk were selective culturing for resistant bacteria, qPCR and shotgun metagenomics
- This data is not yet published, but overall we see relatively good correlation between qPCR and metagenomics. This is not true for all genes, though, and unfortunately neither qPCR nor metagenomics is always better than the other
- Culturing data is not very good at predicting specific antibiotic resistance gene abundances as the class level
- Finally, we have developed methods for discovering new types of ARGs, as seen in the ResFinderFG database: Gschwind et al. 2023
- We have also used these new methods to look at differences between established ARGs and latent ARGs in a variety of environments: Inda-Díaz et al. 2023
- Our ultimate goal in EMBARK would be to develop a modular framework for environmental monitoring of antibiotic resistance. You can read more about our thinking and goals in the review paper we published last year: Bengtsson-Palme et al. 2023
We’re hiring a postdoc in environmental AMR monitoring
As part of the SEARCHER program, we are now hiring a two-year postdoc to work with innovative approaches to antibiotic resistance monitoring. You can read more about the position here and at Chalmers’ job portal, but in short we are after a wet-lab postdoc who are willing to do field work and laboratory studies to identify novel antibiotic resistance genes.
Please do not send me your CV and application letter via e-mail, but apply through the Chalmers application portal. Sending your CV to me will not increase your changes. Only contact me about this position if you have actual, relevant questions about the position (as I will otherwise get lots of unwanted e-mails…) Those questions, I am happy to answer!
Published paper: The latent resistome
What is the latent resistome? This is a term we coin in a new paper published yesterday in Microbiome. In the paper, we distinguish between the small number antibiotic resistance genes (ARGs) that are established, well-characterized, and available in existing resistance gene databases (what we refer to as “established ARGs”). These are typically ARGs encountered in clinical pathogens and are often already causing problems in human and animal infections. The remaining latently present ARGs, which we denote “latent ARGs”, are less or not at all studied, and are therefore much harder to detect (1). These latent ARGs are typically unknown and generally overlooked in most studies of resistance. They are also seldom accounted for in risk assessments of antibiotic resistance (2-4). This means that our view of the resistome and its diversity is incomplete, which hampers our ability to assess risk for promotion and spread of yet undiscovered resistance determinants.
In our new study, we try to alleviate this issue by analyzing more than 10,000 metagenomic samples. We show that the latent ARGs are more abundant and diverse than established ARGs in all studied environments, including the human- and animal-associated microbiomes. The total pan-resistomes, i.e., all ARGs present in an environment (including the latent ARGs), are heavily dominated by these latent ARGs. In contrast, the core resistome (the ARGs that are commonly encountered) comprise both latent and established ARGs.
In the study, we identified several latent ARGs that were shared between environments or that are already present in human pathogens. These are often located on mobile genetic elements that can be transferred between bacteria. Finally, we also show that wastewater microbiomes have surprisingly large pan- and core-resistomes, which makes this environment a potent high-risk environment for mobilization and promotion of latent ARGs, which may make it into pathogens in the future.
It is also interesting to note that this new study echoes the results of my own study from 2018, showing that soil and water environments contain a high diversity of latent ARGs (or ARGs not found in pathogens as I put it in the 2018 study), despite being almost devoid of established ARGs (5).
This project has been a collaboration with Erik Kristiansson’s research group, and particularly with Juan Inda-Diáz. It has been great fun to work with them and I hope that we will keep this collaboration going into the future! The study can be read in its entirety here.
References
- Inda-Díaz JS, Lund D, Parras-Moltó M, Johnning A, Bengtsson-Palme J, Kristiansson E: Latent antibiotic resistance genes are abundant, diverse, and mobile in human, animal, and environmental microbiomes. Microbiome, 11, 44 (2023). doi: 10.1186/s40168-023-01479-0 [Paper link]
- Martinez JL, Coque TM, Baquero F: What is a resistance gene? Ranking risk in resistomes. Nature Reviews Microbiology 2015, 13:116–123. doi:10.1038/nrmicro3399
- Bengtsson-Palme J, Larsson DGJ: Antibiotic resistance genes in the environment: prioritizing risks. Nature Reviews Microbiology, 13, 369 (2015). doi: 10.1038/nrmicro3399-c1
- Bengtsson-Palme J: Assessment and management of risks associated with antibiotic resistance in the environment. In: Roig B, Weiss K, Thoreau V (Eds.) Management of Emerging Public Health Issues and Risks: Multidisciplinary Approaches to the Changing Environment, 243–263. Elsevier, UK (2019). doi: 10.1016/B978-0-12-813290-6.00010-X
- Bengtsson-Palme J: The diversity of uncharacterized antibiotic resistance genes can be predicted from known gene variants – but not always. Microbiome, 6, 125 (2018). doi: 10.1186/s40168-018-0508-2