Metagenomics and the Hype Cycle
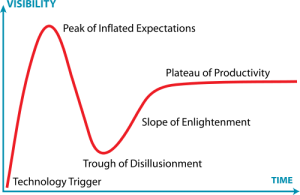
I was creating the diagram below an upcoming presentation, and I realized that the exponential growth in published metagenomics papers might be coming to an end. Interestingly enough the small drop in pace the recent years (701 -> 983 -> 1148) reminds me of the Hype Cycle, where we would (if my projection holds) have reached the “Peak of Inflated Expectations”, which means that we will see a rapid drop in the number of metagenomics publications in the next few years, as the field moves on.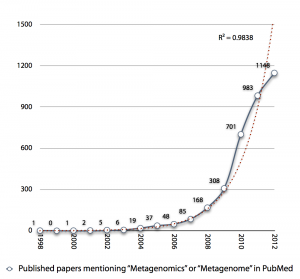
 The thought is interesting, but it seems a little bit early to draw any conclusions from the number of publications, yet. It is still kind of strange to note, though, that more than 20% of metagenomics publications (740/3547) are review papers. Come on, let’s do some science first and then review it… Anyway, it’ll be interesting to see what 2013 has in store for us.
The thought is interesting, but it seems a little bit early to draw any conclusions from the number of publications, yet. It is still kind of strange to note, though, that more than 20% of metagenomics publications (740/3547) are review papers. Come on, let’s do some science first and then review it… Anyway, it’ll be interesting to see what 2013 has in store for us.
Published paper: Guidelines for DNA quality checking
I have co-authored a paper together with, among others, Henrik Nilsson that was published today in MycoKeys. The paper deals with checking quality of DNA sequences prior to using them for research purposes. In our opinion, a lot of the software available for sequence quality management is rather complex and resource intensive. Not everyone have the skills to master such software, and in addition computational resources might also be scarce. Luckily, there’s a lot that can be done in quality control of DNA sequences just using manual means and a web browser. This paper puts these means together into one comprehensible and easy-to-digest document. Our targeted audience is primaily biologists who do not have a strong background in computer science, and still have a dataset requiring DNA sequence quality control.
We have chosen to focus on the fungal ITS barcoding region, but the guidelines should be pretty general and applicable to most groups of organisms. In very short our five guidelines spells:
- Establish that the sequences come from the intended gene or marker
Can be done using a multiple alignment of the sequences and verifying that they all feature some suitable, conserved sub-region (the 5.8S gene in the ITS case) - Establish that all sequences are given in the correct (5’ to 3’) orientation
Examine the alignment for any sequences that do not align at all to the others; re-orient these; re-run the alignment step; and examine them again - Establish that there are no (at least bad cases of) chimeras in the dataset
Run the sequences through BLAST in one of the large sequence databases, e.g. at NCBI (or in the ITS case, use the UNITE database), to verify that the best match comprises more or less the full length of the query sequences - Establish that there are no other major technical errors in the sequences
Examine the BLAST results carefully, particularly the graphical overview and the pairwise alignment, for anomalies (there are some nice figures in the paper on how it should and should not look like) - Establish that any taxonomic annotations given to the sequences make sense
Examine the BLAST hit list to see that the species names produced make sense
A much more thorough description of these guidelines can be found in the paper itself, which is available under open access from MycoKeys. There’s simply no reason not to go there and at least take a look at it. Happy quality control!
Reference
Nilsson RH, Tedersoo L, Abarenkov K, Ryberg M, Kristiansson E, Hartmann M, Schoch CL, Nylander JAA, Bergsten J, Porter TM, Jumpponen A, Vaishampayan P, Ovaskainen O, Hallenberg N, Bengtsson-Palme J, Eriksson KM, Larsson K-H, Larsson E, Kõljalg U: Five simple guidelines for establishing basic authenticity and reliability of newly generated fungal ITS sequences. MycoKeys. Issue 4 (2012), 37–63. doi: 10.3897/mycokeys.4.3606 [Paper link]
Megraft paper in print
I just learned from Research in Microbiology that the paper on our software Megraft has now been assigned a volume and an issue. The proper way of referencing Megraft should consequently now be:
Bengtsson J, Hartmann M, Unterseher M, Vaishampayan P, Abarenkov K, Durso L, Bik EM, Garey JR, Eriksson KM, Nilsson RH: Megraft: A software package to graft ribosomal small subunit (16S/18S) fragments onto full-length sequences for accurate species richness and sequencing depth analysis in pyrosequencing-length metagenomes. Research in Microbiology. Volume 163, Issues 6–7 (2012), 407–412, doi: 10.1016/j.resmic.2012.07.001. [Paper link]
Megraft is currently at version 1.0.1, but I have a slightly updated version in the pipeline which will be made available later this fall.
New paper accepted: Megraft
Yesterday, our paper on Megraft – a software tool to graft ribosomal small subunit (16S/18S) fragments onto full-length SSU sequences – became available as an accepted online early article in Research in Microbiology. Megraft is built upon the notion that when examining the depth of a community sequencing effort, researchers often use rarefaction analysis of the ribosomal small subunit (SSU/16S/18S) gene in a metagenome. However, the SSU sequences in metagenomic libraries generally are present as fragmentary, non-overlapping entries, which poses a great problem for this analysis. Megraft aims to remedy this problem by grafting the input SSU fragments from the metagenome (obtained by e.g. Metaxa) onto full-length SSU sequences. The software also uses a variability model which accounts for observed and unobserved variability. This way, Megraft enables accurate assessment of species richness and sequencing depth in metagenomic datasets.
The algorithm, efficiency and accuracy of Megraft is thoroughly described in the paper. It should be noted that this is not a panacea for species richness estimates in metagenomics, but it is a huge step forward over existing approaches. Megraft shares some similarities with EMIRGE (Miller et al., 2011), which is a software package for reconstruction of full-length ribosomal genes from paired-end Illumina sequences. Megraft, however, is set apart in that it has a strong focus on rarefaction, and functions also when the number of sequences is small, which is often the case in 454 and Sanger-based metagenomics studies. Thus, EMIRGE and Megraft seek to solve a roughly similar problem, but for different sequencing technologies and sequencing scales.
Megraft is available for download here, and the paper can be read here.
-
Bengtsson, J., Hartmann, M., Unterseher, M., Vaishampayan, P., Abarenkov, K., Durso, L., Bik, E.M., Garey, J.R., Eriksson, K.M., Nilsson R.H. (2012). Megraft: A software package to graftribosomal small subunit (16S/18S) fragments onto full-length sequences for accurate species richness and sequencing depth analysis in pyrosequencing-length metagenomes and similar environmental datasets. Research in Microbiology, doi: 10.1016/j.resmic.2012.07.001.
- Miller, C. S., Baker, B. J., Thomas, B. C., Singer, S. W., & Banfield, J. F. (2011). EMIRGE: reconstruction of full-length ribosomal genes from microbial community short read sequencing data. Genome Biology, 12(5), R44. doi:10.1186/gb-2011-12-5-r44
Published paper: Metaxa
It is a pleasure to annonce that the paper on Metaxa is now available as an Online early article in Antonie van Leeuwenhoek. In short, the paper describes a software tool that is able to extract small subunit (SSU) rRNA sequences from large data sets, such as metagenomes and environmental PCR libraries, and classify them according to bacterial, archaeal, eukaryote, chloroplast or mitochondrial origin. The program makes it easy to distinguish between e.g. the bacterial SSU sequences you like to analyze, and the SSU sequences you would like to remove prior to the analysis (e.g. mitochondrial and chloroplast sequences). This task is particularly important in metagenomics, where sequences can potentially derive from a variety of origins, but bacterial diversity often is the desired target for analysis. The software can be downloaded here, and the article can be read here. I would like to thank all the co-authors on this paper for a brilliant collaboration, and hope to be working with them again.
Reference:
- Bengtsson J, Eriksson KM, Hartmann M, Wang Z, Shenoy BD, Grelet G, Abarenkov K, Petri A, Alm Rosenblad M, Nilsson RH: Metaxa: A software tool for automated detection and discrimination among ribosomal small subunit (12S/16S/18S) sequences of archaea, bacteria, eukaryotes, mitochondria, and chloroplasts in metagenomes and environmental sequencing datasets. Antonie van Leeuwenhoek Journal of Microbiology, 2011, doi:10.1007/s10482-011-9598-6.