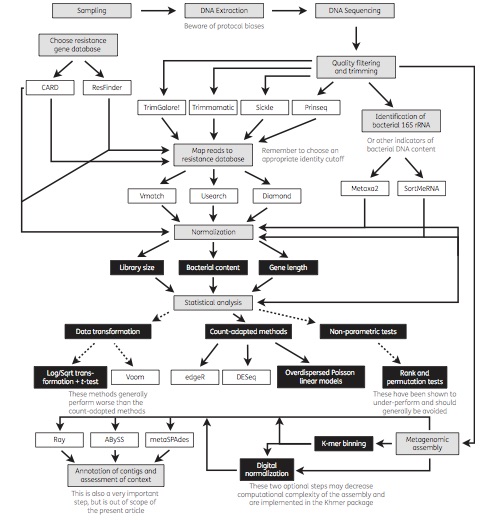
Published paper: Investigating resistomes using metagenomics
Today, a review paper which I wrote together with Joakim Larsson and Erik Kristiansson was published in Journal of Antimicrobial Chemotherapy (1). We have for a long time used metagenomic DNA sequencing to study antibiotic resistance in different environments (2-6), including in the human microbiota (7). Generally, our ultimate purpose has been to assess the risks to human health associated with resistance genes in the environment. However, a multitude of methods exist for metagenomic data analysis, and over the years we have learned that not all methods are suitable for the investigation of resistance genes for this purpose. In our review paper, we describe and discuss current methods for sequence handling, mapping to databases of resistance genes, statistical analysis and metagenomic assembly. We also provide an overview of important considerations related to the analysis of resistance genes, and end by recommending some of the currently used tools, databases and methods that are best equipped to inform research and clinical practice related to antibiotic resistance (see the figure from the paper below). We hope that the paper will be useful to researchers and clinicians interested in using metagenomic sequencing to better understand the resistance genes present in environmental and human-associated microbial communities.
References
- Bengtsson-Palme J, Larsson DGJ, Kristiansson E: Using metagenomics to investigate human and environmental resistomes. Journal of Antimicrobial Chemotherapy, advance access (2017). doi: 10.1093/jac/dkx199 [Paper link]
- Bengtsson-Palme J, Boulund F, Fick J, Kristiansson E, Larsson DGJ: Shotgun metagenomics reveals a wide array of antibiotic resistance genes and mobile elements in a polluted lake in India. Frontiers in Microbiology, 5, 648 (2014). doi: 10.3389/fmicb.2014.00648 [Paper link]
- Lundström S, Östman M, Bengtsson-Palme J, Rutgersson C, Thoudal M, Sircar T, Blanck H, Eriksson KM, Tysklind M, Flach C-F, Larsson DGJ: Minimal selective concentrations of tetracycline in complex aquatic bacterial biofilms. Science of the Total Environment, 553, 587–595 (2016). doi: 10.1016/j.scitotenv.2016.02.103 [Paper link]
- Bengtsson-Palme J, Hammarén R, Pal C, Östman M, Björlenius B, Flach C-F, Kristiansson E, Fick J, Tysklind M, Larsson DGJ: Elucidating selection processes for antibiotic resistance in sewage treatment plants using metagenomics. Science of the Total Environment, 572, 697–712 (2016). doi: 10.1016/j.scitotenv.2016.06.228 [Paper link]
- Pal C, Bengtsson-Palme J, Kristiansson E, Larsson DGJ: The structure and diversity of human, animal and environmental resistomes. Microbiome, 4, 54 (2016). doi: 10.1186/s40168-016-0199-5 [Paper link]
- Flach C-F, Pal C, Svensson CJ, Kristiansson E, Östman M, Bengtsson-Palme J, Tysklind M, Larsson DGJ: Does antifouling paint select for antibiotic resistance? Science of the Total Environment, 590–591, 461–468 (2017). doi: 10.1016/j.scitotenv.2017.01.213 [Paper link]
- Bengtsson-Palme J, Angelin M, Huss M, Kjellqvist S, Kristiansson E, Palmgren H, Larsson DGJ, Johansson A: The human gut microbiome as a transporter of antibiotic resistance genes between continents. Antimicrobial Agents and Chemotherapy, 59, 10, 6551–6560 (2015). doi: 10.1128/AAC.00933-15 [Paper link]
Report on JRC AMR workshop
In March, I attended a workshop on the role of NGS technologies in the coordinated action plan against antimicrobial resistance, organised by JRC in Italy. I was, together with 14 other experts, invited to discuss where and how sequencing can be used to investigate and manage antibiotic resistance. The report from the workshop has just recently been published, and is available here. There will be follow-up activities on this workshop, which I also hope that I will be able to participate in, since this is an important and very interesting pet topic of mine.
Reference
Published paper: Antibiotic resistance in the food supply chain
I am happy to announce that the opinion/review piece I wrote for Current Opinion in Food Science has been published. The paper (1) extends on some of my thoughts on how high-throughput sequencing and metagenomics can aid in risk assessment of antibiotic resistant bacteria that I outlined in my PhD thesis (2), but specifically focuses on the food supply chain and its role in resistance dissemination and selection.
In the paper, I argue for that the food supply chain is a special type of setting in the resistance puzzle, as it not only serves as a connection between environmental habitats for bacteria and humans, but also sometimes presents a substantial selection for resistance, due to use of antibiotics in agri- and aquaculture. International food standards are clear that both selection and dissemination of foodborne resistance should be considered in the risk analysis of food production (3). However, the current main use of DNA sequencing in food safety is whole genome sequencing to delineate which specific strains that are involved in foodborne disease outbreaks, including the resistance factors they may carry (4,5). Further, I argue that while shotgun metagenomics could be used to screen samples for a large number of genes involved in resistance and virulence in the food supply chain, it would at present be very costly and therefore of doubtful benefit to employ in routine screening programs. Still, metagenomics can contribute knowledge that can be used in quantitative risk assessment of antibiotic resistance in the food supply chain.
The entire paper can be read here.
References
- Bengtsson-Palme J: Antibiotic resistance in the food supply chain: Where can sequencing and metagenomics aid risk assessment? Current Opinion in Food Science, in press (2017). doi: 10.1016/j.cofs.2017.01.010 [Paper link]
- Bengtsson-Palme J: Antibiotic resistance in the environment: a contribution from metagenomic studies. Doctoral thesis (medicine), Department of Infectious Diseases, Institute of Biomedicine, Sahlgrenska Academy, University of Gothenburg, 2016. [Link]
- Codex Alimentarius Commission: Guidelines for risk analysis of foodborne antimicrobial resistance. Food and Agriculture Organization of the United Nations & World Health Organization2011. [Link]
- Franz E, Gras LM, Dallman T: Significance of whole genome sequencing for surveillance, source attribution and microbial risk assessment of foodborne pathogens. Current Opinion in Food Science, 8, 74-79 (2016). doi: 10.1016/j.cofs.2016.04.004
- Stasiewicz MJ, Bakker den HC, Wiedmann M: Genomics tools in microbial food safety. Current Opinion in Food Science, 4, 105-110 (2015). doi: 10.1016/j.cofs.2015.06.002
Webinar on Antimicrobial Resistance and the Environment
I will give a short talk on our findings related to antibiotic resistance associated with pharmaceutical production facilities in India at a one-hour webinar arranged by Healthcare Without Harm, taking place on Thursday, November 3rd, 10.00 CET. The webinar will discuss “hot-spot” environments in which antimicrobial resistance can emerge, such as areas in which there are poor pharmaceutical manufacturing practices, where expired or unused drugs are disposed of in an inappropriate way (i.e. by flushing them down the toilet or sink, or disposing them in household rubbish), and areas in which pharmaceuticals are used for aquaculture or agriculture. This is an important aspect of the resistance problem, but to date most of the actions taken to tackle the spread of AMR don’t take into account this aspect of antimicrobials released into the environment. The webinar is co-organised by HCWH Europe and HCWH Asia, and aims to raise awareness about the issue of AMR and its environmental impact. It features, apart from myself, Lucas Wiarda (Global Marketing Director & Head of Sustainable Antibiotics Program at DSM Sinochem Pharmaceuticals) and Sister Mercilyn Jabel (Pharmacist at Saint Paul Hospital Cavite, Philippines).
Sign up here to learn about:
- Antibiotic pollution and waste
- Recent findings from India regarding antibiotic discharges in rivers from manufacturers and new mechanisms by which resistance spreads in the environment
- Sustainable antibiotics – how to support the proper and effective use of antibiotics and their responsible production
- How the pharmaceutical industry is addressing the environmental pollution that leads to AMR
- The best practices in managing infectious waste at hospital level
Published opinion piece: Why limit antibiotic pollution?
Me and Joakim Larsson wrote an opinion/summary piece for the APUA Newsletter, issued by the Alliance for Prudent Use of Antibiotics, that was published yesterday (1). The paper is essentially a summary of work included in my PhD thesis, and discusses how to establish minimal selective concentrations of antibiotics for microbial communities (2-4), how to identify risk environments for resistance selection (5-9), and which mitigation strategies that can be implemented (10-12). Partially, we also discussed these issues earlier in our paper in the Medicine Maker (10), but this paper goes deeper into why limiting antibiotic pollution is important to mitigate the accelerating antibiotic resistance problem. I recommend this short summary piece to anyone who would like a brief overview of our research on antibiotic resistance, and think that it can serve as a great starting point for further reading! In addition, this issue of the newsletter features very interesting pieces on reducing antibiotics use (and disposal) outside of the clinics (13) and revival of old antibiotics (14). Please go ahead to the APUA web site and read the entire newsletter!
References
- Bengtsson-Palme J, Larsson DGJ: Why limit antibiotic pollution? The role of environmental selection in antibiotic resistance development. APUA Newsletter, 34, 2, 6-9 (2016). [Paper link].
- Bengtsson-Palme J, Larsson DGJ: Concentrations of antibiotics predicted to select for resistant bacteria: Proposed limits for environmental regulation. Environment International, 86, 140-149 (2016). doi: 10.1016/j.envint.2015.10.015 [Paper link]
- Gullberg E, Cao S, Berg OG, Ilbäck C, Sandegren L, Hughes D, et al.: Selection of resistant bacteria at very low antibiotic concentrations. PLoS Pathogens 7, e1002158 (2011).
- Lundström S, Östman M, Bengtsson-Palme J, Rutgersson C, Thoudal M, Sircar T, Blanck H, Eriksson KM, Tysklind M, Flach C-F, Larsson DGJ: Minimal selective concentrations of tetracycline in complex aquatic bacterial biofilms. Science of the Total Environment, 553, 587–595 (2016). doi: 10.1016/j.scitotenv.2016.02.103
- Bengtsson-Palme J, Boulund F, Fick J, Kristiansson E, Larsson DGJ: Shotgun metagenomics reveals a wide array of antibiotic resistance genes and mobile elements in a polluted lake in India. Frontiers in Microbiology, 5, 648 (2014). doi: 10.3389/fmicb.2014.00648
- Bengtsson-Palme J, Hammarén R, Pal C, Östman M, Björlenius B, Flach C-F, Kristiansson E, Fick J, Tysklind M, Larsson DGJ: Elucidating selection processes for antibiotic resistance in sewage treatment plants using metagenomics. Science of the Total Environment, in press (2016). doi: 10.1016/j.scitotenv.2016.06.228
- Berendonk TU, Manaia CM, Merlin C, Fatta-Kassinos D, Cytryn E, Walsh F, et al.: Tackling antibiotic resistance: the environmental framework. Nature Reviews Microbiology, 13, 310–317 (2015). doi: 10.1038/nrmicro3439
- Martinez JL, Coque TM, Baquero F: What is a resistance gene? Ranking risk in resistomes. Nature Reviews Microbiology 2015, 13:116–123. doi:10.1038/nrmicro3399
- Bengtsson-Palme J, Larsson DGJ: Antibiotic resistance genes in the environment: prioritizing risks. Nature Reviews Microbiology, 13, 369 (2015) doi:10.1038/nrmicro3399‐c1
- Bengtsson-Palme J, Larsson DGJ: Time to limit antibiotic pollution. The Medicine Maker, 0416, 302, 17–18 (2016). [Paper link]
- Ashbolt NJ, Amézquita A, Backhaus T, Borriello P, Brandt KK, Collignon P, et al.: Human Health Risk Assessment (HHRA) for Environmental Development and Transfer of Antibiotic Resistance. Environmental Health Perspectives, 121, 993–1001 (2013)
- Pruden A, Larsson DGJ, Amézquita A, Collignon P, Brandt KK, Graham DW, et al.: Management options for reducing the release of antibiotics and antibiotic resistance genes to the environment. Environmental Health Perspectives, 121, 878–85 (2013).
- Theuretzbacher U: Optimizing the Use of Old Antibiotics — A Global Health Agenda. APUA Newsletter, 34, 2, 10-13 (2016). [Paper link].
- Amábile-Cuevas CF: Antibiotics and Antibiotic Resistance All Around Us. APUA Newsletter, 34, 2, 3-5 (2016). [Paper link].
Published paper: Antibiotic resistance in sewage treatment plants
 After a long wait (1), Science of the Total Environment has finally decided to make our paper on selection of antibiotic resistance genes in sewage treatment plants (STPs) available (2). STPs are often suggested to be “hotspots” for emergence and dissemination of antibiotic-resistant bacteria (3-6). However, we actually do not know if the selection pressures within STPs, that can be caused either by residual antibiotics or other co-selective agents, are sufficiently large to specifically promote resistance. To better understand this, we used shotgun metagenomic sequencing of samples from different steps of the treatment process (incoming water, treated water, primary sludge, recirculated sludge and digested sludge) in three Swedish STPs in the Stockholm area to characterize the frequencies of resistance genes to antibiotics, biocides and metal, as well as mobile genetic elements and taxonomic composition. In parallel, we also measured concentrations of antibiotics, biocides and metals.
After a long wait (1), Science of the Total Environment has finally decided to make our paper on selection of antibiotic resistance genes in sewage treatment plants (STPs) available (2). STPs are often suggested to be “hotspots” for emergence and dissemination of antibiotic-resistant bacteria (3-6). However, we actually do not know if the selection pressures within STPs, that can be caused either by residual antibiotics or other co-selective agents, are sufficiently large to specifically promote resistance. To better understand this, we used shotgun metagenomic sequencing of samples from different steps of the treatment process (incoming water, treated water, primary sludge, recirculated sludge and digested sludge) in three Swedish STPs in the Stockholm area to characterize the frequencies of resistance genes to antibiotics, biocides and metal, as well as mobile genetic elements and taxonomic composition. In parallel, we also measured concentrations of antibiotics, biocides and metals.
We found that only the concentrations of tetracycline and ciprofloxacin in the influent water were above those that we predict to cause resistance selection (7). However, there was no consistent enrichment of resistance genes to any particular class of antibiotics in the STPs, neither for biocide and metal resistance genes. Instead, the most substantial change of the bacterial communities compared to human feces (sampled from Swedes in another study of ours (8)) occurred already in the sewage pipes, and was manifested by a strong shift from obligate to facultative anaerobes. Through the treatment process, resistance genes against antibiotics, biocides and metals were not reduced to the same extent as fecal bacteria were.
Worryingly, the OXA-48 beta-lactamase gene was consistently enriched in surplus and digested sludge. OXA-48 is still rare in Swedish clinical isolates (9), but provides resistance to carbapenems, one of our most critically important classes of antibiotics. However, taken together metagenomic sequencing did not provide clear support for any specific selection of antibiotic resistance. Rather, since stronger selective forces affect gross taxonomic composition, and thereby also resistance gene abundances, it is very hard to interpret the metagenomic data from a risk-for-selection perspective. We therefore think that comprehensive analyses of resistant vs. non-resistant strains within relevant species are warranted.
Taken together, the main take-home messages of the paper (2) are:
- There were no apparent evidence for direct selection of resistance genes by antibiotics or co-selection by biocides or metals
- Abiotic factors (mostly oxygen availability) strongly shape taxonomy and seems to be driving changes of resistance genes
- Metagenomic and/or PCR-based community studies may not be sufficiently sensitive to detect selection effects, as important shifts towards resistant may occur within species and not on the community level
- The concentrations of antibiotics, biocides and metals were overall reduced, but not removed in STPs. Incoming concentrations of antibiotics in Swedish STPs are generally low
- Resistance genes are overall reduced through the treatment process, but far from eliminated
References and notes
- Okay, those who takes notes know that I have already complained once before on Science of the Total Environment’s ridiculously long production handling times. But, seriously, how can a journal’s production team return the proofs for after three days of acceptance, and then wait seven weeks before putting the final proofs online? I still wonder what is going on beyond the scenes, which is totally obscure because the production office also refuses to respond to e-mails. Not a nice publishing experience this time either.
- Bengtsson-Palme J, Hammarén R, Pal C, Östman M, Björlenius B, Flach C-F, Kristiansson E, Fick J, Tysklind M, Larsson DGJ: Elucidating selection processes for antibiotic resistance in sewage treatment plants using metagenomics. Science of the Total Environment, in press (2016). doi: 10.1016/j.scitotenv.2016.06.228 [Paper link]
- Rizzo L, Manaia C, Merlin C, Schwartz T, Dagot C, Ploy MC, Michael I, Fatta-Kassinos D: Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: a review. Science of the Total Environment, 447, 345–360 (2013). doi: 10.1016/j.scitotenv.2013.01.032
- Laht M, Karkman A, Voolaid V, Ritz C, Tenson T, Virta M, Kisand V: Abundances of Tetracycline, Sulphonamide and Beta-Lactam Antibiotic Resistance Genes in Conventional Wastewater Treatment Plants (WWTPs) with Different Waste Load. PLoS ONE, 9, e103705 (2014). doi: 10.1371/journal.pone.0103705
- Yang Y, Li B, Zou S, Fang HHP, Zhang T: Fate of antibiotic resistance genes in sewage treatment plant revealed by metagenomic approach. Water Research, 62, 97–106 (2014). doi: 10.1016/j.watres.2014.05.019
- Berendonk TU, Manaia CM, Merlin C, Fatta-Kassinos D, Cytryn E, Walsh F, et al.: Tackling antibiotic resistance: the environmental framework. Nature Reviews Microbiology, 13, 310–317 (2015). doi: 10.1038/nrmicro3439
- Bengtsson-Palme J, Larsson DGJ: Concentrations of antibiotics predicted to select for resistant bacteria: Proposed limits for environmental regulation. Environment International, 86, 140–149 (2016). doi: 10.1016/j.envint.2015.10.015
- Bengtsson-Palme J, Angelin M, Huss M, Kjellqvist S, Kristiansson E, Palmgren H, Larsson DGJ, Johansson A: The human gut microbiome as a transporter of antibiotic resistance genes between continents. Antimicrobial Agents and Chemotherapy, 59, 10, 6551–6560 (2015). doi: 10.1128/AAC.00933-15
- Hellman J, Aspevall O, Bengtsson B, Pringle M: SWEDRES-SVARM 2014. Consumption of antimicrobials and occurrence of antimicrobial resistance in Sweden. Public Health Agency of Sweden and National Veterinary Institute, Solna/Uppsala, Sweden. Report No.: 14027. Available from: http://www.folkhalsomyndigheten.se/publicerat-material/ (2014)
Environmental pollution with antibiotics leads to resistance
So, on Thursday (May 26th) I will defend my thesis, titled “Antibiotic resistance in the environment: a contribution from metagenomic studies”. I will not dwell into this by writing a novel text, but will instead shamelessly reproduce the press release, which should give a reasonable overview of what I have been doing:
More and more people are infected with antibiotic resistant bacteria. But how do bacteria become resistant? A doctoral thesis from the Centre for Antibiotic Resistance Research at University of Gothenburg has investigated the role of the environment in the development of antibiotic resistance.
“An important question we asked was how low concentrations of antibiotics that can favour the growth of resistant bacteria in the environment”, says Johan Bengtsson-Palme, author of the thesis.
“Based on our analyses, we propose emission limits for 111 antibiotics that should not be exceeded in order to avoid that environmental bacteria become more resistant.”
A starting point to regulate antibiotic pollution
A recent report, commissioned by the British Prime Minister David Cameron, proposes that the emission limits suggested in Johan’s thesis should be used as a starting point to regulate antibiotic pollution from, for example, pharmaceutical production – globally.
“Many people are surprised that such regulations are not already in place, but today it is actually not a crime to discharge wastewater contaminated with large amounts of antibiotics, not even in Europe”, says Johan Bengtsson-Palme.
Resistance genes
In one of the studies in the thesis, the researchers show that resistance genes against a vast range of antibiotics are enriched in an Indian lake polluted by dumping of wastewater from pharmaceutical production.
“It’s scary. Not only do the bacteria carry a multitude of resistance genes. They are also unusually well adapted to share those genes with other bacteria. If a disease-causing bacterium ends up in the lake, it may quickly pick up the genes it needs to become resistant. Since the lake is located close to residential areas, such spread of resistant bacteria to humans is not hard to imagine”, says Johan Bengtsson-Palme.
Spreading by travelers
The thesis also shows that resistant bacteria spread in the intestines of travelers who have visited India or Central Africa, even if the travelers themselves have not become sick.
“That resistant bacteria spread so quickly across the planet highlights that we must adopt a global perspective on the resistance problem”, says Johan Bengtsson-Palme. “Furthermore, it is not enough to reduce the use of antibiotics in healthcare. We must also reduce the use of antibiotics for animals, and try to limit the releases of antibiotics into the environment to try to get control over the growing antibiotic resistance problem before it is too late”.
The thesis Antibiotic resistance in the environment: a contribution from metagenomic studies will be defended on a dissertation on May 26th.
Published opinion piece: Time to limit antibiotic pollution
In the most recent issue of the Medicine Maker (#0416), there is a short opinion piece by me and Joakim Larsson, in which we argue for that pharmaceutical companies should live up to their ethical responsibilities, and may actually benefit from doing so (1). We were invited to write for the Medicine Maker based on our recent papers on proposed limits for antibiotic discharges into the environment (2) and minimal selective concentrations (3).
We argue that now as PNECs for resistance selection are available, they should be applied in regulatory contexts. The recent O’Neill report on antimicrobial resistance (commissioned by the British Government) specifically highlighted the urgent need for enforceable regulations on antibiotic discharges (4). The concentrations we reported in our Environment International paper (2) can be used by local, national and international authorities to define emission limits for antibiotic-producing factories, but also for pharmaceutical companies to assess and manage risks for resistance selection associated with their own discharges.
The Medicine Maker can be read for free, but requires registration to access its full content. The full opinion piece can be found here.
References
- Bengtsson-Palme J, Larsson DGJ: Time to limit antibiotic pollution. The Medicine Maker, 0416, 302, 17–18 (2016). [Paper link]
- Bengtsson-Palme J, Larsson DGJ: Concentrations of antibiotics predicted to select for resistant bacteria: Proposed limits for environmental regulation. Environment International, 86, 140–149 (2016). doi: 10.1016/j.envint.2015.10.015 [Paper link]
- Lundström S, Östman M, Bengtsson-Palme J, Rutgersson C, Thoudal M, Sircar T, Blanck H, Eriksson KM, Tysklind M, Flach C-F, Larsson DGJ: Minimal selective concentrations of tetracycline in complex aquatic bacterial biofilms. Science of the Total Environment, 553, 587–595 (2016). doi: 10.1016/j.scitotenv.2016.02.103 [Paper link]
- Review on Antimicrobial Resistance: Antimicrobials in agriculture and the environment: Reducing unnecessary use and waste (J O’Neill, Ed,) (2015). [Link].
Published paper: Community MSCs for tetracycline
After a long wait (1) Sara Lundström’s paper establishing minimal selective concentrations (MSCs) for the antibiotic tetracycline in complex microbial communities (2), of which I am a co-author, has gone online. Personally, I think this paper is among the finest work I have been involved in; a lot of good science have gone into this publication. Risk assessment and management of antibiotics pollution is in great need of scientific data to underpin mitigation efforts (3). This paper describes a method to determine the minimal selective concentrations of antibiotics, and investigates different endpoints for measuring those MSCs. The method involves a testing system highly relevant for aquatic communities, in which bacteria are allowed to form biofilms in aquaria under controlled antibiotic exposure. Using the system, we find that 1 μg/L tetracycline selects for the resistance genes tetA and tetG, while 10 μg/L tetracycline is required to detect changes of phenotypic resistance. In short, the different endpoints studied (and their corresponding MSCs) were:
- CFU counts on R2A plates with 20 μg/mL tetracycline – MSC = 10 μg/L
- MIC range – MSC ~ 10-100 μg/L
- PICT, leucine uptake after short-term TC challenge – MSC ~ 100 μg/L
- Increased resistance gene abundances, metagenomics – MSC range: 0.1-10 μg/L
- Increased resistance gene abundances, qPCR (tetA and tetG) – MSC ≤ 1 μg/L
- Changes to taxonomic diversity – no significant changes detected
- Changes to taxonomic community composition – MSC ~ 1-10 μg/L
This study confirms that the estimated PNECs we reported recently (4) correspond well to experimentally determined MSCs, at least for tetracycline. Importantly, the selective concentrations we report for tetracycline overlap with those that have been reported in sewage treatment plants (5). We also see that tetracycline not only selects for tetracycline resistance genes, but also resistance genes against other classes of antibiotics, including sulfonamides, beta-lactams and aminoglycosides. Finally, the approach we describe can be used for improved in risk assessment for (also other) antibiotics, and to refine the emission limits we suggested in a recent paper based on theoretical calculations (4).
References and notes
- Okay, seriously: how can a journal’s production team return the proofs for a paper within 24 hours of acceptance, and then wait literally five weeks before putting the final proofs online? Nothing against STOTEN, but I honestly wonder what was going on beyond the scenes here.
- Lundström SV, Östman M, Bengtsson-Palme J, Rutgersson C, Thoudal M, Sircar T, Blanck H, Eriksson KM, Tysklind M, Flach C-F, Larsson DGJ: Minimal selective concentrations of tetracycline in complex aquatic bacterial biofilms. Science of the Total Environment, 553, 587–595 (2016). doi: 10.1016/j.scitotenv.2016.02.103 [Paper link]
- Ågerstrand M, Berg C, Björlenius B, Breitholtz M, Brunstrom B, Fick J, Gunnarsson L, Larsson DGJ, Sumpter JP, Tysklind M, Rudén C: Improving environmental risk assessment of human pharmaceuticals. Environmental Science and Technology (2015). doi:10.1021/acs.est.5b00302
- Bengtsson-Palme J, Larsson DGJ: Concentrations of antibiotics predicted to select for resistant bacteria: Proposed limits for environmental regulation. Environment International, 86, 140-149 (2016). doi: 10.1016/j.envint.2015.10.015
- Michael I, Rizzo L, McArdell CS, Manaia CM, Merlin C, Schwartz T, Dagot C, Fatta-Kassinos D: Urban wastewater treatment plants as hotspots for the release of antibiotics in the environment: a review. Water Research, 47, 957–995 (2013). doi:10.1016/j.watres.2012.11.027
Published paper: Predicted selective concentrations for antibiotics
Yesterday was an intensive day for typesetters apparently, since they put two of my papers online on the same day. This second paper was published in Environment International, and focuses on predicting minimal selective concentrations for all antibiotics present in the EUCAST database (1).
Today (well, up until yesterday at least), we have virtually no knowledge of which environmental concentrations that can exert a selection pressure for antibiotic resistant bacteria. However, experimentally determining minimal selective concentrations (MSCs) in complex ecosystems would involve immense efforts if done for all antibiotics. Therefore, efforts to theoretically determine MSCs for different antibiotics have been suggested (2,3). In this paper we therefore estimate upper boundaries for selective concentrations for all antibiotics in the EUCAST database, based on the assumption that selective concentrations a priori must be lower than those completely inhibiting growth. Data on Minimal Inhibitory Concentrations (MICs) were obtained for 122 antibiotics and antibiotics combinations, the lowest observed MICs were identified for each of those across all tested species, and to compensate for limited species coverage, we adjusted the lowest MICs for the number of tested species. We finally assessed Predicted No Effect Concentrations (PNECs) for resistance selection using an assessment factor of 10 to account for the differences between MICs and MSCs. Since we found that the link between taxonomic similarity between species and lowest MIC was weak, we have not compensated for the taxonomic diversity that each antibiotic was tested against – only for limited number of species tested. In most cases, our PNECs for selection of resistance were below available PNECs for ecotoxicological effects retrieved from FASS. Also, concentrations predicted to be selective have, for some antibiotics, been detected in regular sewage treatment plants (4), and are greatly exceeded in environments polluted by pharmaceutical pollution (5-7), often with drastic consequences in terms of resistance gene enrichments (8-10). This is a central issue since in principle a transfer event of a novel resistance determinant from an environmental bacteria to an (opportunistic) human pathogen only need to occur once to become a clinical problem (11). Once established, the gene could then spread through human activities, such as trade and travel (7,13). Importantly, this paper:
- Provides upper boundaries for selective concentrations (MSCs) for 111 antibiotics
- Predicts no effect concentrations (PNECs) for resistance selection
- Can guide implementation of compound-specific emission limits based on the provided concentrations
The paper is available under open access here. We hope, and believe, that the data will be of great use in environmental risk assessments, in efforts by industries, regulatory agencies or purchasers of medicines to define acceptable environmental emissions of antibiotics, in the implementation of environmental monitoring programs, for directing mitigations, and for prioritizing future studies on environmental antibiotic resistance.
References:
- Bengtsson-Palme J, Larsson DGJ: Concentrations of antibiotics predicted to select for resistant bacteria: Proposed limits for environmental regulation. Environment International, 86, 140-149 (2016). doi: 10.1016/j.envint.2015.10.015 [Paper link]
- Ågerstrand M, Berg C, Björlenius B, Breitholtz M, Brunstrom B, Fick J, Gunnarsson L, Larsson DGJ, Sumpter JP, Tysklind M, Rudén C: Improving environmental risk assessment of human pharmaceuticals. Environmental Science and Technology (2015). doi:10.1021/acs.est.5b00302
- Tello A, Austin B, Telfer TC: Selective pressure of antibiotic pollution on bacteria of importance to public health. Environmental Health Perspectives, 120, 1100–1106 (2012). doi:10.1289/ehp.1104650
- Michael I, Rizzo L, McArdell CS, Manaia CM, Merlin C, Schwartz T, Dagot C, Fatta-Kassinos D: Urban wastewater treatment plants as hotspots for the release of antibiotics in the environment: a review. Water Research, 47, 957–995 (2013). doi:10.1016/j.watres.2012.11.027
- Larsson DGJ, de Pedro C, Paxeus N: Effluent from drug manufactures contains extremely high levels of pharmaceuticals. Journal of Hazardous Materials, 148, 751–755 (2007). doi:10.1016/j.jhazmat.2007.07.008
- Fick J, Söderström H, Lindberg RH, Phan C, Tysklind M, Larsson DGJ: Contamination of surface, ground, and drinking water from pharmaceutical production. Environmental Toxicology and Chemistry, 28, 2522–2527 (2009). doi:10.1897/09-073.1
- Larsson DGJ: Pollution from drug manufacturing: review and perspectives. Philosophical Transactions of the Royal Society London, Series B Biological Sciences, 369 (2014). doi:10.1098/rstb.2013.0571
- Bengtsson-Palme J, Boulund F, Fick J, Kristiansson E, Larsson DGJ: Shotgun metagenomics reveals a wide array of antibiotic resistance genes and mobile elements in a polluted lake in India. Frontiers in Microbiology, Volume 5, Issue 648 (2014). doi: 10.3389/fmicb.2014.00648 [Paper link]
- Kristiansson E, Fick J, Janzon A, Grabic R, Rutgersson C, Weijdegård B, Söderström H, Larsson DGJ: Pyrosequencing of antibiotic-contaminated river sediments reveals high levels of resistance and gene transfer elements. PLoS ONE, Volume 6, e17038 (2011). doi:10.1371/journal.pone.0017038.
- Marathe NP, Regina VR, Walujkar SA, Charan SS, Moore ERB, Larsson DGJ, Shouche YS: A Treatment Plant Receiving Waste Water from Multiple Bulk Drug Manufacturers Is a Reservoir for Highly Multi-Drug Resistant Integron-Bearing Bacteria. PLoS ONE, Volume 8, e77310 (2013). doi:10.1371/journal.pone.0077310
- Bengtsson-Palme J, Larsson DGJ: Antibiotic resistance genes in the environment: prioritizing risks. Nature Reviews Microbiology, 13, 369 (2015). doi: 10.1038/nrmicro3399-c1 [Paper link]
- Bengtsson-Palme J, Angelin M, Huss M, Kjellqvist S, Kristiansson E, Palmgren H, Larsson DGJ, Johansson A: The human gut microbiome as a transporter of antibiotic resistance genes between continents. Antimicrobial Agents and Chemotherapy, 59, 10, 6551-6560 (2015). doi: 10.1128/AAC.00933-15 [Paper link]