My ISME talk on EMBARK
Ákos Kovács had the brilliant idea of putting up a temporary resource for things you bring up in a talk that you can point people to. I did not do this before my talk at ISME today, but I thought the idea was so good, so here’s a summary and collection of my ISME short-talk on the EMBARK outcomes today:
- More information on EMBARK and its successor SEARCHER can be found on the project website, here: http://antimicrobialresistance.eu Importantly, this is a team effort over four years and I only touched on a few selected things
- Within the project we have looked at typical background levels of antibiotic resistance in the environment. We have already published some of these results (for qPCR abundances) in Abramova et al. 2023
- The average resistance gene in the average environment is present in ~1 in 1000 bacteria, but the variation between different genes is huge
- Depending on monitoring goal, different target genes are relevant to use. See this table adapted from Abramova et al. 2023:
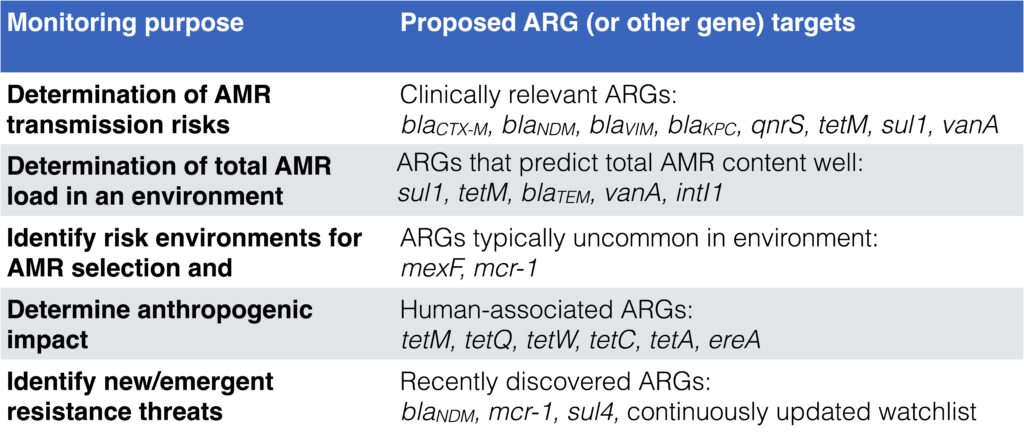
- We have also tried to make different monitoring methods for environmental AMR comparable. Those mentioned in the talk were selective culturing for resistant bacteria, qPCR and shotgun metagenomics
- This data is not yet published, but overall we see relatively good correlation between qPCR and metagenomics. This is not true for all genes, though, and unfortunately neither qPCR nor metagenomics is always better than the other
- Culturing data is not very good at predicting specific antibiotic resistance gene abundances as the class level
- Finally, we have developed methods for discovering new types of ARGs, as seen in the ResFinderFG database: Gschwind et al. 2023
- We have also used these new methods to look at differences between established ARGs and latent ARGs in a variety of environments: Inda-Díaz et al. 2023
- Our ultimate goal in EMBARK would be to develop a modular framework for environmental monitoring of antibiotic resistance. You can read more about our thinking and goals in the review paper we published last year: Bengtsson-Palme et al. 2023
Open positions!
First of all, I just want to do a last reminder of PhD student position in bioinformatics and artificial intelligence applied to antibiotic resistance with Erik Kristiansson as main supervisor that closes tomorrow. More info here!
Second, two of my best and dearest colleagues at University of Gothenburg – Kaisa Thorell and Åsa Sjöling – have open postdoc positions in molecular microbiology (with Åsa) and bacterial proteomics (with Kaisa). Both of these are great opportunities to work with fantastic people on exciting subjects, so you should check these out if you are looking for postdoc positions in microbiology, molecular biology or bioinformatics! There are only a few days left to apply for these positions, so go ahead and do it now!
Finally, I am again tooting our own horn with the postdoc in innovative approaches to antibiotic resistance monitoring (within the SEARCHER program) in my own group. More info here, deadline is on July 31 with interviews to take place in August.
We’re hiring a postdoc in environmental AMR monitoring
As part of the SEARCHER program, we are now hiring a two-year postdoc to work with innovative approaches to antibiotic resistance monitoring. You can read more about the position here and at Chalmers’ job portal, but in short we are after a wet-lab postdoc who are willing to do field work and laboratory studies to identify novel antibiotic resistance genes.
Please do not send me your CV and application letter via e-mail, but apply through the Chalmers application portal. Sending your CV to me will not increase your changes. Only contact me about this position if you have actual, relevant questions about the position (as I will otherwise get lots of unwanted e-mails…) Those questions, I am happy to answer!