BIG NEWS: We’re moving to Chalmers
I have very big and exciting news to share with you. After more than 10 years at the Sahlgrenska Academy, me and my lab will be moving from the University of Gothenburg to Chalmers University of Technology (which is physically a move of less than a kilometer, so still within Gothenburg). I have been offered a position at the Division of Systems Biology, funded by the SciLifeLab and Wallenberg National Program for Data-Driven Life Science (DDLS). The total funding to my lab will be 17 million SEK, with some co-funding from Chalmers added in on top of that.
I am of course very excited about this opportunity, which will bring some infrastructure that we need in-house that we don’t have easy access to today. At the same time, I am sad to leave my academic ‘home’, and the fantastic people we have been working with there for the years. I am also endlessly thankful for the support and trust that the Sahlgrenska Academy, the Institute of Biomedicine and the Department of Infectious Diseases have put into me and my research over the past years.
The transition to Chalmers will start already in May, but will be gradual and continue for a long time. We have close ties to the Sahlgrenska Academy and we will keep closely collaborating with researchers there. I will also retain an affiliation to the University of Gothenburg, at least for the near future.
All in all, this year will bring very interesting development, and this additional funding from the DDLS program will allow us to venture into new areas of bioinformatics and try out ideas that have previously been out of reach. I look forward to work with our new colleagues at Chalmers and within the DDLS program in the coming years!
Swedish Bioinformatics Workshop 2014
I am part of the organizing committee for the Swedish Bioinformatics Workshop (#SBW2014) that will be held October 23-24 this year in Gothenburg. I would like to invite you all, especially master/PhD students and PostDocs in Sweden, to come and share the event with us!
SBW is an annual event that has been organized by the different universities in Sweden. This year it will take place at the Wallenberg Conference Centre in Gothenburg and is arranged by both University of Gothenburg and Chalmers University of Technology. SBW2014 will, as the tradition abides, be a meeting point for PhD students and postdocs working with any kind of bioinformatics within Sweden and is therefore free of charge for these groups. We are proud to announce a program including both invited speakers – such as Mick Watson from the Roslin institute, Dawn Field from University of Oxford, and Joakim Lundeberg from KTH – along with participant presentations and poster sessions. This year, the program will also contain a number of workshop sessions where hands-on problems will be used as starting points for discussions on new bioinformatics approaches to these problems. This will provide opportunities for attendees with different methodological backgrounds to interact and work together to find synergies between fields and come up with creative solutions.
More information about the event including registration and abstract submission can be found at www.sbw2014.se.
I, and the rest of the organizers, look forward to meeting you in Gothenburg in October!
Webpage: http://www.sbw2014.se
Facebook: https://www.facebook.com/events/1450513325188910/
Google+: https://plus.google.com/events/cuhlpovcc275stut854dk5ussnk
If you want, you can spread the word, for example using this flyer!
Swedish Bioinformatics Workshop 2011
The 11th annual meeting of PhD students and Postdoc researchers in Bioinformatics in Sweden will take place in Lund the 29-30 September. The workshop is an opportunity for young researchers to meet, exchange ideas, and keep up to date with the growing body of knowledge. I will go there, and you should be there too! Besides, it’s free for PhD students and Postdocs! All info can be found at the workshop website. The last time I was there (2010) really fueled some interesting discussions, and I am really looking forward to the event this year. Hope to meet you there, fellow Swedish bioinformaticians!
Get ready for the future: Microbial Community Systems Biology
Phil Goetz at JCVI recently posted his reflections from the Summit of Systems Biology. I was not there, but I read his summary with interest. Now, what strikes me as interesting is the notion that “there were no talks on metagenomics. This also struck me as odd; bacterial communities seem like a natural systems biology problem.” Having been working with microbial communities for a while, I am surprised that the modeling perspective that is so prevalent in macro-organism ecosystems ecology have not yet really come to fruition in microbial ecology. With the tremendous amounts of sequences that are pouring over us from microbial communities, and with the plethora of functional metagenomics annotation that is made, how come that there has been so little research in the actual interactions between microorganisms within e.g. biofilms?
The problem is also connected to the lack of time-series data from community research. To be able to understand how a system behaves under changing conditions, we need to measure its reactions to various parameter changes over time. Instead of pooling metagenomes to reduce temporal “noise” we need to be better at identifying the changing parameters and then use the temporal differences to look for responses to the parameter changes. By applying a functional metagenomics perspective at each sample point, combining this with measured changes in community species structure (as measured e.g. by 16S or some other marker gene), and correlating this with changes in the parameters, we should be able to build a model of how the ecosystem responds to changing environments. With the large-scale sequencing technologies available today, and the possibilities given by metatranscriptomics, these ideas should be challenging but not impossible.
I am not saying that any of these things have not been done. But it has been done to a surprisingly small extent. I would highly appreciate reading a paper trying to build a mathematical model of how the ecosystem functions in bacterial communities shift in response to an environmental stressor. Because when someone builds such a model we suddenly have a tool to take microbial community research from an explorative perspective to an applied one. The applied perspective will be useful for actually protecting environments and ecosystem services, as well as for understanding how to manipulate microbial ecosystems to maximize the outtake beneficial to society. Also, the understanding the ecosystem dynamics of microbial systems could be carried over to macro-ecosystems and provide a small-scale ecosystem laboratory for all ecosystem research. Such a shift towards applied microbial community systems biology will be more or less necessary to be able to argue for more resources and time being spent on e.g. metagenomics. And I believe that we will soon be there, because the step is shorter than we might imagine.
Thesis presentation
I will present my master thesis “Metagenomic Analysis of Marine Periphyton Communities”, on Tuesday the 22nd of March, at 13.00. The presentation will take place in the room Folke Andreasson at Medicinaregatan 11 in Gothenburg. The presentation is open for everyone, but the number of seats are limited.
BLAST Visualization
Perhaps because of my roots in systems biology (or the cause of going there in the first place), I have always had an interest in creating visually appealing images of data, many times in the form of networks. I find that often in bioinformatics, one of the hardest problems is to make information understandable. For example, a BLAST output might say very little about how the genes or proteins are connected to each other, at least to the untrained eye.
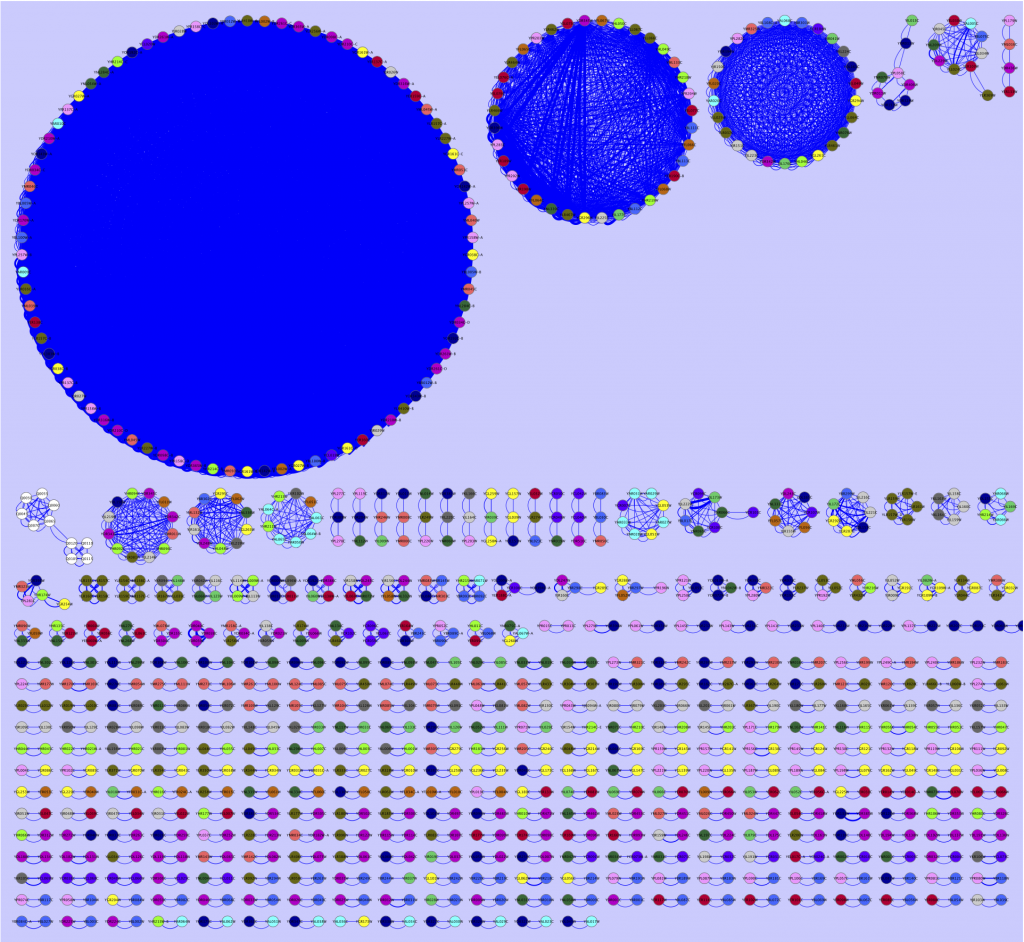
Therefore, during the last weeks I have fiddled around with various ways of viewing interesting portions of BLAST reports. By making all-against-all BLAST searches, and outputting the data in table format (blastall option -m 8), I have been able to extract the hits I am interested in and export them into a Cytoscape compatible format, with some accompanying metadata (scores, e-values, alignment length, etc.). The results are many times pretty unparsable by the eye, rendering them a bit meaningless, but have been more and more interesting as I have put more effort into the extraction script. Just as an example, I here provide a simple map of the best all-against-all matches in the Saccharomyces cerevisiae genome, as a Cytoscape network (click for full size):
The largest circle consists of transposable elements (jumping DNA which inserts itself at multiple locations in the genome, no surprise there is a lot of them, and that these are pretty conserved). The circle to the left of the transposon circle consists of genes located inside the telomeric regions. Why they show such high similarity I do not know, but it seems plausible that the telomere thing could play a role here. The third circle contain mostly members of the seripauperin multigene family, which is also located close to the telomeres. At the bottom you found the gene pairs, that match to each other. You could go on with all the smaller structures as well, but I am no yeast expert, so I will stop here, letting this serve as an example of what a BLAST report really look like.
For this image, I have used a blastn report of all yeast ORFs (taken from yeastgenome.org) as input to my extraction tool, selected Cytoscape compatible output, and used a maximal e-value of 0.00001 and an alignment length of at least 50 nts as criteria to be extracted. I have also pooled the sequences according to chromosome number. The pooling was used to color code the nodes in Cytoscape. The edge width is connected to alignment score, a high score renders a thick line, and a low score causes the line to be thin.
I am still working on the extraction tool and will not provide any code yet. Input would, however, be appreciated. My personal opinion is that in the near future, the overload of newly produced DNA and protein sequences will choke us if do not come up with more intuitive ways of displaying data. I don’t think that the network above is there yet. Still, it conveys information I would not have been able to understand from just looking at the BLAST output. The first attempts to come around the sequence overload problem won’t be the best ones. But we got to start working on visualization methods today, so that we do not end up with sequences over our shoulders in just a few years. Besides, a network image seems much more impressive than a number of lines of text…
FEBS Workshop: Last Chance
The time is running out if you want to attend to the workshop session on mapping signal transduction, hosted by Stefan Hohmann and Marcus Krantz, which I will take part in. Deadline is on the 15 of May, so register soon if you have not already done. You can find all important info here.
The workshop will take place on June 29:th, between 13.00 and 15.30. The goal is to show some visualisation strategies for signal transduction pathways, and how to use pathway maps as a base to create mathematical models. There will be a brief introduction to mapping and modelling and to the software used (Cytoscape, CellDesigner). This will be followed by independent work with a set of small case studies that demonstrates the basic methodology. I will take part in answering questions and assisting during the case study part.