Get ready for the future: Microbial Community Systems Biology
Phil Goetz at JCVI recently posted his reflections from the Summit of Systems Biology. I was not there, but I read his summary with interest. Now, what strikes me as interesting is the notion that “there were no talks on metagenomics. This also struck me as odd; bacterial communities seem like a natural systems biology problem.” Having been working with microbial communities for a while, I am surprised that the modeling perspective that is so prevalent in macro-organism ecosystems ecology have not yet really come to fruition in microbial ecology. With the tremendous amounts of sequences that are pouring over us from microbial communities, and with the plethora of functional metagenomics annotation that is made, how come that there has been so little research in the actual interactions between microorganisms within e.g. biofilms?
The problem is also connected to the lack of time-series data from community research. To be able to understand how a system behaves under changing conditions, we need to measure its reactions to various parameter changes over time. Instead of pooling metagenomes to reduce temporal “noise” we need to be better at identifying the changing parameters and then use the temporal differences to look for responses to the parameter changes. By applying a functional metagenomics perspective at each sample point, combining this with measured changes in community species structure (as measured e.g. by 16S or some other marker gene), and correlating this with changes in the parameters, we should be able to build a model of how the ecosystem responds to changing environments. With the large-scale sequencing technologies available today, and the possibilities given by metatranscriptomics, these ideas should be challenging but not impossible.
I am not saying that any of these things have not been done. But it has been done to a surprisingly small extent. I would highly appreciate reading a paper trying to build a mathematical model of how the ecosystem functions in bacterial communities shift in response to an environmental stressor. Because when someone builds such a model we suddenly have a tool to take microbial community research from an explorative perspective to an applied one. The applied perspective will be useful for actually protecting environments and ecosystem services, as well as for understanding how to manipulate microbial ecosystems to maximize the outtake beneficial to society. Also, the understanding the ecosystem dynamics of microbial systems could be carried over to macro-ecosystems and provide a small-scale ecosystem laboratory for all ecosystem research. Such a shift towards applied microbial community systems biology will be more or less necessary to be able to argue for more resources and time being spent on e.g. metagenomics. And I believe that we will soon be there, because the step is shorter than we might imagine.
2x+ Metaxa speedup on the way
I’m working on an update to Metaxa that will bring at least double speed to the program (and even more when run on really large data sets on many cores). While there is still no real release version of this update (version 1.1), I have today posted a public “beta”, which you can use for testing purposes. Do not use this version for anything important (e.g. research) as it contains at least one known bug (and maybe even more I haven’t discovered yet). I would appreciate, if you are interested, that you download this version and e-mailed any bugs or inconsistencies found to me (firstname.lastname[at]microbiology.se).
Note that to install this version, you first need to download and install the current version of Metaxa (1.0.2). Then the new version can be used with the old’s databases.
Using Metaxa to automatically classify SSUs to the species level
One potential use for Metaxa (paper) is to include it in a pipeline for classification of SSU rRNA in metagenomic data (or other environmental sequencing sets). However, as Metaxa is provided from this site, it only classifies SSUs to the domain level (archaea, bacteria and eukaryotes, with the addition of chloroplasts and mitochondria). It is also able to do some (pretty rough) species guesses using the “--guess_species T” option. An easy solution to implement would be to pass the Metaxa output, e.g. “metaxa_output.bacteria.fasta” to BLAST, and compare all these sequences to the sequences in e.g. the SILVA or GreenGenes database. There is, however, a way to improve this, which uses Metaxa’s ability to compares sequences to custom databases. In this tutorial, I will show you how to achieve this.
Before we start, you will of course need to download and install Metaxa, and its required software packages (BLAST, HMMER, MAFFT). When you have done this, we can get going with the database customization. I will in this tutorial use the SILVA database for SSU classification. However, the basic idea for the tutorial should be easily applicable to GreenGenes and other rRNA databases as well.
- Visit SILVA through this link, and download the file named “SSURef_106_tax_silva.fasta.tgz”. The file is pretty big so it may take a while to download it. If you’re running Metaxa on a server, you’ll have to get the SILVA-file to the server somehow.
- Unzip and untar the file (Mac OS X makes this neatly by doubleclicking the file, on linux you can do it on the command line by typing “
tar -xvzf SSURef_106_tax_silva.fasta.tgz“). This will give you a FASTA-file. - The FASTA-file needs to be prepared a bit for Metaxa usage. First, we need to give Metaxa identifiers it can understand. Metaxa identifies sequences’ origins by the last character in their identifier, e.g. “>A16379.1.1496.B”. Here, “.B” indicates that this is a bacterial sequence. We are now going to use the unix command sed to process the file and insert the appropriate identifiers.
- We begin with the archaeal sequences. To get those straight, we type:
sed "s/ Archaea;/.A - Archaea;/" SSURef_106_tax_silva.fasta > temp1
Notice that we direct the output to a temporary file. It is bad practice to replace the input file with the output file, so we work with two temp-files instead. - The next step is also easy, now we find all eukaryote sequences and add E:s to the identifiers:
sed "s/ Eukaryota;/.E - Eukaryota;/" temp1 > temp2 - Now it becomes a little more complicated, as SILVA classes mitochondrial and chloroplast SSU sequences as subclasses of bacteria. However, there is a neat little trick we can use. First we do the same with the bacterial sequences as with the archaeal and eukaryote:
sed "s/ Bacteria;/.B - Bacteria;/" temp2 > temp1 - Now, we can use two a little more complicated commands to annotate the mitochondrial and chloroplast sequences:
sed "s/\.B - \(Bacteria;.*;[Mm]itochondria;\)/.M - \1/" temp1 > temp2
sed "s/\.B - \(Bacteria;.*;[Cc]hloroplast;\)/.C - \1/" temp2 > temp1 - We also need to get “rid” of the unclassified sequences, by assigning them to the “other” origin (O):
sed "s/ Unclassified;/.O - Unclassified;/" temp1 > temp2
- We begin with the archaeal sequences. To get those straight, we type:
- That wasn’t too complicated, was it? We can now check the number of different sequences in the file by typing the pretty complicated command:
grep ">" temp2 | cut -f 1 -d " " | rev | cut -f 1 -d "." | sort | uniq -c
If you have been working with the same files as me, you should now see the following numbers:
23172 A
471949 B
3712 C
55937 E
534 M
226 O - At this stage, we need to remove the full taxonomy from the FASTA headers, as Metaxa cannot handle species names of this length. We do this by typing:
sed "s/ - .*;/ - /" temp2 > temp1 - We can now change the temp-file into a FASTA file, and delete the other temp-file:
mv temp1 SSURef.fasta
rm temp2 - We now need to configure Metaxa to use the database. First, we format a BLAST-database from the FASTA-file we just created:
formatdb -i SSURef.fasta -t "SSURef Metaxa DB" -o T -p F - With that done, we can now run Metaxa using this database instead of the classification database that comes with the program. By specifying that we want to guess the species origin of sequences, we can get (as accurate as SILVA lets us be) which species each sequence in our set come from. We do this by using the
-dand the--guess_speciesoptions:
metaxa -i test.fasta -d SSURef.fasta -o TEST --guess_species T --cpu 2
The input in this case was the test file that comes with Metaxa. Note also that we’re using two CPUs to get multithreaded speeds. Remember that you must provide the full (or relative) path to the database files we just created, if you are not running Metaxa from the same directory as the database resides in. - The output should now look like this (taken from the bacterial file):
>coryGlut_Bielefeld_dna Bacterial 16S SSU rRNA, best species guess: Corynebacterium glutamicum
CGAACGCTG...
>gi|116668568:792344-793860 Bacterial 16S SSU rRNA, best species guess: Arthrobacter sp. J3.40
TGAACGCTG...
>gi|117927211:c1399163-1397655 Bacterial 16S SSU rRNA, best species guess: Acidothermus cellulolyticus
>CGAACGCTG...
And so on. As you can see the species names are now located at the end of each definition line, and can easily be extracted using sed, e.g. “grep ">" TEST.bacteria.fasta | sed "s/.*: //"“.
And that’s it. It’s pretty simple, and can easily be scripted. In fact, I have already made the bash script for you. That means that the short version is, download the script, download the sequence file from SILVA, move into the directory you have downloaded the file to and run the script by typing: ./prepare_silva_for_metaxa.sh
A few notes at the end. The benefit of using this approach is that we maintain the sorting capabilities, marking of uncertain sequences and error checking of Metaxa, but we don’t have to add another BLAST step after Metaxa has finished. However, as this database we create is a lot bigger than the database that comes with Metaxa, the running time of the classification step will be substantially longer. This is in most cases acceptable, as that time is the same as the time it would have taken to run BLAST on the Metaxa output. It should also be noted that this approach limits Metaxa’s ability of classifying 12S sequences, as there are no such sequences in SILVA. Good luck with classifying your metagenome SSUs (and if you use Metaxa in your research, remember to cite the paper)!
Antibiotic resistance driving virulence?
It seriously worries me that a number of indications recently have pointed to that the heavy use of antibiotics does not only drive antibiotic resistance development, but also the development towards more virulent and aggressive strains of pathogenic bacteria. First, the genome sequencing of the E. coli strain that caused the EHEC outbreak in Germany in May revealed not only antibiotic resistance genes, but also is also able to make Shiga toxin, which is causes the severe diarrhoea and kidney damage related to the haemolytic uremic syndrome (HUS). The genes encoding the Shiga toxin are not originally bacterial genes, but instead seem to originate from phages. When E. coli gets infected with a Shiga toxin-producing phage, it becomes a human pathogen [1]. David Acheson, managing director for food safety at consulting firm Leavitt Partners, says that exposure to antibiotics might be enhancing the spread of Shiga toxin-producing phage. Some antibiotics triggers what is referred to as the SOS response, which induces the phage to start replicating. The replication of the phage causes the bacteria to burst, releasing the phages, and with them the toxin [1].
Second, there is apparently an ongoing outbreak of scarlet fever in Hong Kong. Kwok-Yung Yuen, microbiologist at the University of Hong Kong, has analyzed the draft sequence of the genome, and suggests that the bacteria acquired greater virulence and drug resistance by picking up one or more genes from bacteria in the human oral and urogenital tracts. He believes that the overuse of antibiotics is driving the emergence of drug resistance in these bacteria [2].
Now, both of these cases are just indications, but if they are true that would be an alarming development, where the use of antibiotics promotes the spread not only of resistance genes, impairing our ability to treat bacterial infections, but also the development of far more virulent and aggressive strains. Combining increasing untreatability with increasing aggressiveness seems to me like the ultimate weapon against our relatively high standards of treatment of common infections. Good thing hand hygiene still seems to help [3].
References
- Phage on the rampage (http://www.nature.com/news/2011/110609/full/news.2011.360.html), Published online 9 June 2011, Nature, doi:10.1038/news.2011.360
- Mutated Bacteria Drives Scarlet Fever Outbreak (http://news.sciencemag.org/scienceinsider/2011/06/mutated-bacteria-drives-scarlet.html?etoc&elq=cd94aa347dca45b3a82f144b8213e82b), Published online 27 June 2011.
- Luby SP, Halder AK, Huda T, Unicomb L, Johnston RB (2011) The Effect of Handwashing at Recommended Times with Water Alone and With Soap on Child Diarrhea in Rural Bangladesh: An Observational Study. PLoS Med 8(6): e1001052. doi:10.1371/journal.pmed.1001052 (http://www.plosmedicine.org/article/info%3Adoi%2F10.1371%2Fjournal.pmed.1001052)
Questions? Suggestions?
So Metaxa has gone into the wild, which means that I start to get feedback from users using it in ways I have not foreseen. This is the best and the worst thing about having your software exposed to real-world usage; it makes it possible to improve it in a variety of ways, but it also gives you severe headaches at times. I could luckily fix a smaller bug in the Metaxa code within a matter of hours and issue an update to version 1.0.2. The interesting thing here was that I would never have discovered the bug myself, as I never would have called the Metaxa program in the way required for the bug to happen. But once I saw the command given, and the output, which the user kindly sent me, I pretty quickly realized what was wrong, and how to fix it. Therefore, I would like to ask all out you who use Metaxa to send me your questions, problems and bug reports. The feedback is highly appreciated, and I can (at least currently) promise to issue fixes as fast as possible. We are really committed to make Metaxa work for everyone.
If you have suggestions for improvements, those are welcome as well (though it will take significantly more time to implement new features than to fix bugs). I am currently compiling a FAQ, and all questions are welcome. Finally, I would like to thank everybody who has downloaded and tried the Metaxa package. I can see in the server logs that there are quite many of you, which of course makes us happy.
From the literature batch…
A random sample of things from this week’s scientific news I think are worth sharing:
Britain is apparently shutting down many of its climate change outreach efforts. I find this very saddening, and see it as an indication of our extreme short-sightedness. We need to put more effort and funding into preserving the environment – not less. In addition, the economic benefits of taking care of the nature around us will probably be much larger than the small sums we save in the short term by not doing anything. We clearly need better incentives to look beyond the next budget and the next election.
The editorial of Nature Reviews Microbiology points the torch on the need for research within basic microbiology, pointing out that “the functions of many genes in the genomes of even the best studied organisms, such as Escherichia coli and Bacillus subtilis, remain unknown. Often these genes do not resemble other, characterized, genes in the databases, allowing for the possibility that interesting new pathways remain to be discovered. (…) if we want to understand how life works at the molecular level, it is crucial to continue and expand basic microbiology research.” I would like to add that a more complete understanding of at least one model organism would drastically increase the accuracy of genome (and metagenome) annotation in new sequencing projects, which today is patchy, to say the least.
Moving on…
So, last week I started my Ph.D. in Joakim Larsson’s group at the Sahlgrenska Academy. While I am very happy about how things have evolved, I will also miss the ecotox group and the functional genomics group a lot (though both do their research within 10 minutes walking distance from my new place…) I spent last week getting through the usual administrative hassle; getting keys and cards, signing papers, installing bioinformatics software on my new monster of a computer etc. Slowly, the new room starts to feel like it is mine (after nailing phylogenetic trees, my favorite map of the amino acids, and my remember-why-Cytoscape-visualisation-might-not-be-a-good-idea-for-all-network-like-structures poster to the billboard).
So what will this change of positions mean? Will I quit doing research on microbial communities? Of course not! In my new position, my subject of investigation will be bacterial communities subjected to antibiotics. We will look for resistance genes in such communities, and try to answer questions like: How do a high antibiotic selection pressure affect abundance of resistance genes and mobile elements that could facilitate their transfer between bacteria? Can resistance genes found in environmental bacteria be transferred to the microbes of the human gut? Can the environmental bacteria tell us what resistance genes that will be present in clinical situations in the near future? All these questions could, at least partially, be answered by metagenomic approaches and good bioinformatics tools, and my role will be to come up with the solutions provide answers to them.
I am excited over this new project, which involves my favorite subject – metagenomics and community analysis – as well as important factors, such as the clinical connections, the possibility to add pieces to the antibiotic resistance puzzle, and the role of gene and species transfer in resistance development. I also like the fact that I will need to handle high-throughput sequence data, meaning that there will be many opportunities to develop tools, a task I highly enjoy. I think the next couple of years will be an exciting time.
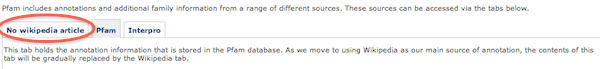
Pfam + Wikipedia – finally!
Browsing the Pfam web site today, I discovered that the database finally has launched its Wikipedia co-ordination efforts.
This has happened along with the 25th release of the Pfam database (released 1st of April), and basically means that Wikipedia articles will be linked to Pfam families. Gradually, this will (hopefully) improve the annotation of Pfam families, which has in many cases been rather poor. The Xfam blog post related to Pfam release 25 says the change will be happening gradually, which might actually be good thing, given the quirks that might pop up.
(…) a major change is that Pfam annotation is now beginning to be co-ordinated via Wikipedia. Unlike Rfam, where every entry has a Wikipedia entry, we expect this to be a more gradual transition for Pfam, so not all entries currently have a corresponding Wikipedia article. For a more detailed discussion, check the help page. We actively encourage the addition of new/updated annotations via Wikipedia as they will appear far quicker than waiting for a Pfam release. If there are articles in Wikipedia that you think correspond to a family, then please mail us!
I have awaited this change for a long time, and is very happy that Pfam has finally taken this step. Congratulations and my sincerest thanks to the Pfam team! Now, let’s go editing!
Thesis presentation
I will present my master thesis “Metagenomic Analysis of Marine Periphyton Communities”, on Tuesday the 22nd of March, at 13.00. The presentation will take place in the room Folke Andreasson at Medicinaregatan 11 in Gothenburg. The presentation is open for everyone, but the number of seats are limited.
Underpinning Wikipedia’s Wisdom
In December, Alex Bateman, whose opinions on open science I support and have touched upon earlier, wrote a short correspondence letter to Nature [1] in which he again repeated the points of his talk at FEBS last summer. He concludes by the paragraph:
Many in the scientific community will admit to using Wikipedia occasionally, yet few have contributed content. For society’s sake, scientists must overcome their reluctance to embrace this resource.
I agree with this statement. However, as I also touched upon earlier, but like to repeat again – bold statements doesn’t make dreams come true – action does. Rfam, and the collaboration with RNA Biology and Wikipedia is a great example of such actions. So what other actions may be necessary to get researchers to contribute to the Wikipedian wisdom?
First of all, I do not think that the main obstacle to get researchers to edit Wikipedia articles is reluctance to doing so because Wikipedia is “inconsistent with traditional academic scholarship”, though that might be a partial explanation. What I think is the major problem is the time-reward tradeoff. Given the focus on publishing peer-reviewed articles, the race for higher impact factor, and the general tendency of measuring science by statistical measures, it should be no surprise that Wikipedia editing is far down on most scientists to-do lists, so also on mine. The reward of editing a Wikipedia article is a good feeling in your stomach that you have benefitted society. Good stomach feelings will, however, feed my children just as little as freedom of speech. Still, both Wikipedia editing and freedom of speech are extremely important, especially as a scientist.
Thus, there is a great need of a system that:
- Provides a reward or acknowledgement for Wikipedia editing.
- Makes Wikipedia editing economically sustainable.
- Encourages publishing of Wikipedia articles, or contributions to existing ones as part of the scientific publishing process.
Such a system could include a “contribution factor” similar to the impact factor, in which contribution of Wikipedia and other open access forums was weighted, with or without a usefulness measure. Such a usefulness measure could easily be determined by links from other Wikipedia articles, or similar. I realise that there would be severe drawbacks of such a system, similar to those of the impact factor system. I am not a huge fan of impact factors (read e.g. Per Seglen’s 1997 BMJ article [2] for some reasons why), but I do not see that system changing any time soon, and thus some kind of contribution factor could provide an additional statistical measure for evaluators to consider when examining scientists’ work.
While a contribution factor would be an incitement for researchers to contribute to the common knowledge, it will still not provide an economic value to do so. This could easily be changed by allowing, and maybe even requiring, scientists to contribute to Wikipedia and other public fora of scientific information as part of their science outreach duties. In fact, this public outreach duty (“tredje uppgiften” in Swedish) is governed in Swedish law. In 2009, the universities in Sweden have been assigned to “collaborate with the society and inform about their operations, and act such that scientific results produced at the university benefits society” (my translation). It seems rational that Wikipedia editing would be part of that duty, as that is the place were many (most?) people find information online today. Consequently, it is only up to the universities to demand 30 minutes of Wikipedia editing per week/month from their employees. Note here that I am referring to paid editing.
Another way of increasing the economic appeal of writing Wikipedia articles would be to encourage funding agencies and foundations to demand Wikipedia articles or similar as part of project reports. This would require researchers to make their findings public in order to get further funding, a move that would greatly increase the importance of increasing the common wisdom treasure. However, I suspect that many funding agencies, as well as researchers would be reluctant to such a solution.
Lastly, as shown by the Rfam/RNA Biology/Wikipedia relationship, scientific publishing itself could be tied to Wikipedia editing. This process could be started by e.g. open access journals such as PLoS ONE, either by demanding short Wikipedia notes to get an article published, or by simply provide prioritised publishing of articles which also have an accompanying Wiki-article. As mentioned previously, these short Wikipedia notes would also go through a peer-review process along with the full article. By tying this to the contribution factor, further incitements could be provided to get scientific progress in the hands of the general public.
Now, all these ideas put a huge burden on already hard-working scientists. I realise that they cannot all be introduced simultaneously. Opening up publishing requires time and thought, and should be done in small steps. But doing so is in the interest of scientists, the general public and the funders, as well as politicians. Because in the long run it will be hard to argue that society should pay for science when scientists are reluctant to even provide the public with an understandable version of the results. Instead of digging such a hole for ourselves, we should adapt the reward, evaluation, funding and publishing systems in a way that they benefit both researchers and the society we often say we serve.
- Bateman and Logan. Time to underpin Wikipedia wisdom. Nature (2010) vol. 468 (7325) pp. 765
- Seglen. Why the impact factor of journals should not be used for evaluating research. BMJ (1997) vol. 314 (7079) pp. 498-502