Mumame – Quantifying mutations in metagenomes
Let me get straight to something somewhat besides the point here: summer students can achieve amazing things! One such student I had the pleasure to work with this summer is Shruthi Magesh, and a preprint based on work she did with me at the Wisconsin Institute for Discovery this summer just got published on bioRxiv (1). The preprint describes a software tool called Mumame, which uses database information on mutations in DNA or protein sequences to search metagenomic datasets and quantifies the relative proportion of resistance mutations over wild type sequences.
In the preprint (1), we first of all show that Mumame works on amplicon data where we already knew the true outcome (2). Second, we show that we can detect differences in mutation frequencies in controlled experiments (2,3). Lastly, we use the tool to gain some further information about resistance patterns in sediments from polluted environments in India (4,5). Together these analyses show that one of the most central aspects for Mumame to be able to find mutations is having a very high number of sequenced reads in all libraries (preferably more than 50 million per library), because these mutations are generally rare – even in polluted environments and microcosms exposed to antibiotics. We expect Mumame to be a useful addition to metagenomic studies of e.g. antibiotic resistance, and to increase the detail by which metagenomes can be screened for phenotypically important differences.
While I did write the code for the software (with a lot of input from Viktor Jonsson, who also is a coauthor on the preprint, on the statistical analysis), Shruthi did the software testing and evaluations, and the paper would not have been possible hadn’t she wanted a bioinformatic summer project related to metagenomics, aside from her laboratory work. The resulting preprint is available from bioRxiv and the Mumame software is freely available from this site.
References
- Magesh S, Jonsson V, Bengtsson-Palme J: Quantifying point-mutations in metagenomic data. bioRxiv, 438572 (2018). doi: 10.1101/438572 [Link]
- Kraupner N, Ebmeyer S, Bengtsson-Palme J, Fick J, Kristiansson E, Flach C-F, Larsson DGJ: Selective concentration for ciprofloxacin in Escherichia coli grown in complex aquatic bacterial biofilms. Environment International, 116, 255–268 (2018). doi: 10.1016/j.envint.2018.04.029 [Paper link]
- Lundström S, Östman M, Bengtsson-Palme J, Rutgersson C, Thoudal M, Sircar T, Blanck H, Eriksson KM, Tysklind M, Flach C-F, Larsson DGJ: Minimal selective concentrations of tetracycline in complex aquatic bacterial biofilms. Science of the Total Environment, 553, 587–595 (2016). doi: 10.1016/j.scitotenv.2016.02.103 [Paper link]
- Bengtsson-Palme J, Boulund F, Fick J, Kristiansson E, Larsson DGJ: Shotgun metagenomics reveals a wide array of antibiotic resistance genes and mobile elements in a polluted lake in India. Frontiers in Microbiology, 5, 648 (2014). doi: 10.3389/fmicb.2014.00648 [Paper link]
- Kristiansson E, Fick J, Janzon A, Grabic R, Rutgersson C, Weijdegård B, Söderström H, Larsson DGJ: Pyrosequencing of antibiotic-contaminated river sediments reveals high levels of resistance and gene transfer elements. PLoS ONE, Volume 6, e17038 (2011). doi:10.1371/journal.pone.0017038.
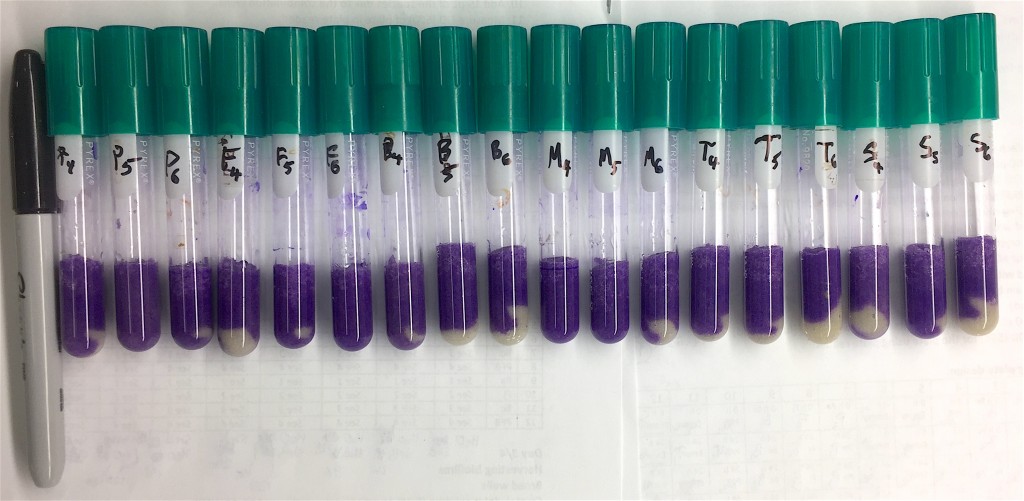
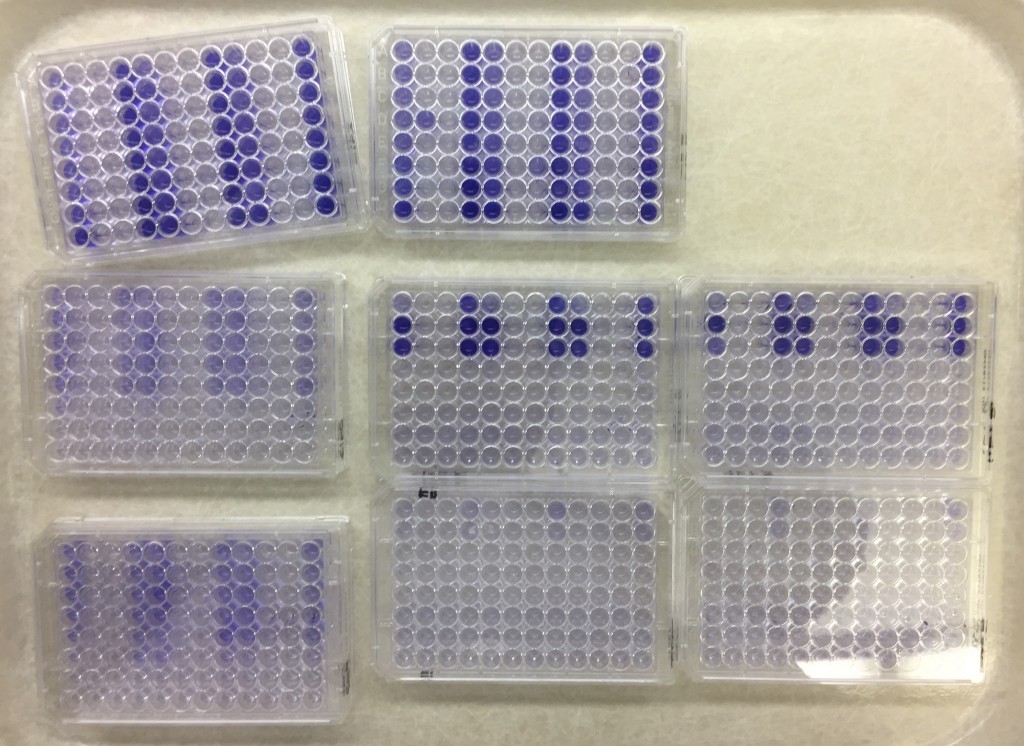
Can you spot the difference?
Can you spot the differences in color shade in this picture?
Me neither. And that’s why I suspect that I’ve spent 30 hours or so on something completely not useful stuff. I hereby take weekend. See you next week!
The Wisconsin Blog is on again
I’ve been having a very intense start of the year with the move to the US and getting the family accustomed to Madison (which has taken time and energy, but gone really well). I just wanted to make you aware of that I have started posting at the Wisconsin Blog again and hope to be sharing research related stuff from my year in the US there. For more personal stuff, our family has set up a blog (in Swedish) at this address: https://palmeiamerikat.blogspot.com. You are very welcome to follow our adventure there!
And the experiments have started!
It’s been a long time before I have written something here, mostly because making ourselves at home in Madison have take some time; then we go the flue; and then there have been a lot at work after that. But now i will try to have another go at writing somethings on the Wisconsin Blog. First, a look at my (already messy) desk here at the Wisconsin Institute for Discovery.
And then, a look at my lab space, which I have a view of straight from my desk, through a glass window.
This week, I have started experiments with exposing our little model community to antibiotics and it looks like I’m getting potentially exciting results. I have to sit down with the data today to see if there’s statistical differences, but from the looks of the biofilms, there is potential here.
Next week I will try to start experiment with sand columns and see if I can replicate some of this in this setting as well. It is interesting being back in the lab, and I feel that this an experience that will be very valuable for me going forward. I look forward to the days later this spring when I will start generating sequence data from my own experiments!
Leaving Madison




Something to live up to




Around the biochemistry building of the University of Wisconsin-Madison campus (and perhaps also elsewhere), there are a lot of these signs, highlighting discoveries that were made in Madison. And they really have a history to be proud of. This place is where both bacterial conjugation and transduction was discovered, paving the way for much of the genome editing and studies we take for granted today. And they also did the basics required to generate knock-out mice for genetics studies. I could go on, but I I think I made my point. This place has a lost of history. And I doubt that my slightly messy project will live up to these groundbreaking discoveries.
Labor day

Before that, I went to see a house, which I liked a lot, so hopefully I will be able to sign a contract for this before I leave for Sweden again on Wednesday. Fingers crossed, over and out!
Farmers Market



The rest of the day I spent walking around by the Lake Monona and Lake Wingra, taking a break for a coffee at Colectivo Coffee. Very nice place with excellent pourovers. But now my feet hurts and I need to get some sleep!




Students are coming back




Manuel, my hero
Okay, first of all this is a shoutout post to Manuel Garavito, who has been putting up with me for the last two days – my first two days in a wet lab for several years. Manuel has been beyond fantastic in showing me their basic protocols and having patience with my rusty lab skills. If this project will work out, it will very much be because of him.
 This weekend, there haven’t been a lot of time for other things than lab work, but I spent Friday evening at the Memorial Union where I happened to stumble into a concert with Brazilian (I think, not sure) music. And on Saturday evening I went out to the Hop Cat, where I tasted the fantastic beer Psychedellic Cat Grass and got taught the basics of American football by a woman who was also there on her own, apparently to watch the game. So, I’m doing my well but working my ass off with things I am not really that good at. Yet.
This weekend, there haven’t been a lot of time for other things than lab work, but I spent Friday evening at the Memorial Union where I happened to stumble into a concert with Brazilian (I think, not sure) music. And on Saturday evening I went out to the Hop Cat, where I tasted the fantastic beer Psychedellic Cat Grass and got taught the basics of American football by a woman who was also there on her own, apparently to watch the game. So, I’m doing my well but working my ass off with things I am not really that good at. Yet.