ITSx moved out of beta
As promised yesterday, I have now uploaded an update to ITSx, bringing it to version 1.0.2. So what’s new in this version?
First of all, ITSx is now taken out of beta and is now considered ready for production use. We do no longer find any bugs in it, and since there’s now a wide range of people already using it for various purposes, we feel confident that any significant bugs would have been unraveled by now.
Secondly, I have also added support for the new HMMER version (3.1b) released in May in this version of ITSx. So you can now go ahead and install HMMER 3.1 if you want to try out the new HMMER beta and still be able to use ITSx.
Finally, we have also updated the manual somewhat, hopefully making it a little easier to use ITSx for a first-time user.
Version 1.0.2 of ITSx can be downloaded from here. As previously, you may still report any bugs, strange behaviors, ideas for new features, or inconsistencies with certain lineages, by mailing to “itsx” at this domain name.
Metaxa and HMMER 3.1b
As you might be aware, a new version of HMMER is out since late May. You might wonder how Metaxa (relying on HMMER3) will work if you update to the new version of HMMER, and I have finally got around to test it! The answer, according to my somewhat limited testing, is that Metaxa 1.1.2 seems to be working fine with HMMER 3.1.
You might need to go into the database directory (“metaxa_db”; should be located in the same directory as the Metaxa binaries), and remove all the files ending with suffixes .h3f .h3i .h3m and .h3p inside the “HMMs” directory. On most installation, this should not be necessary. Myself, I just plugged HMMER 3.1 in and started Metaxa, but if you get error messages complaining that “Error: bad format, binary auxfiles,
binary auxfiles are in an outdated HMMER format (3/b); please hmmpress your HMM file again”, then you should try removing the files and re-running Metaxa. This might especially be a problem on older Metaxa versions. [Update: Note that this fix will likely not work with ITSx!]
Bear in mind that I have not run thorough testing on Metaxa and HMMER 3.1, and probably won’t for the 1.1.2 version, since there’s a 2.0 version waiting just around the corner…
Additionally, if you experience problems with Megraft, you should try the same fix as for Metaxa, but with the Megraft database directory instead. Regarding ITSx, a minor update will be released very soon, which also will address HMMER 3.1b compatibility. [Update: See this post for how to work around HMMER 3.1 problems with ITSx.]
Happy barcoding everyone!
I have joined ResearchGate
On a side note, I just joined Research Gate (my profile). I’ve noted that it generates kind of the same kind of belonging-to-a-group feeling that registering on Facebook did way back, when co-author after co-author starts following you. Still, I haven’t figured out exactly what to use it for yet; it certainly seem more useful than academia.edu, with abilities to ask questions etc., but is anyone of you really using ResearchGate for this? Or is it rather just another showcasing window for researchers (much like my Publications page)? Please feel free do add your opinions as comments to this post!
ITSx – a software tool for detection and extraction of ITS1 and ITS2 sequences
For a couple of years, I have been working with microbial ecology and diversity, and how such features can be assessed using molecular barcodes, such as the SSU (16S/18S) rRNA sequence (the Metaxa and Megraft packages). However, I have also been aiming at the ITS region, and how that can be used in barcoding (see e.g. the guidelines we published last year). It is therefore a great pleasure to introduce my next gem for community analysis; a software tool for detection and extraction of the ITS1 and ITS2 regions of ITS sequences from environmental communities. The tool is dubbed ITSx, and supersedes the more specific fungal ITS extractor written by Henrik Nilsson and colleagues. Henrik is once more the mastermind behind this completely rewritten version, in which I have done the lion’s share of the programming. Among the new features in ITSx are:
- Robust support for the Cantharellus, Craterellus, and Tulasnella genera of fungi
- Support for nineteen additional eukaryotic groups on top of the already present support for fungi (specifically these groups: Tracheophyta (vascular plants), Bryophyta (bryophytes), Marchantiophyta (liverworts), Chlorophyta (green algae), Rhodophyta (red algae), Phaeophyceae (brown algae), Metazoa (metazoans), Oomycota (oomycetes), Alveolata (alveolates), Amoebozoa (amoebozoans), Euglenozoa, Rhizaria, Bacillariophyta (diatoms), Eustigmatophyceae (eustigmatophytes), Raphidophyceae (raphidophytes), Synurophyceae (synurids), Haptophyceae (haptophytes) , Apusozoa, and Parabasalia (parabasalids))
- Multi-processor support
- Extensive output options
- Virtually zero false-positive extractions
ITSx is today moved from a private pre-release state to a public beta state. No code changes has been made since February, indicative of that the last pre-release candidate is now ready to fly on its own. As far as our testing has revealed, this version seems to be bug free. In reality though, researchers tend to find the most unexpected usage scenarios. So please, if you find any unexpected behavior in this version of ITSx, send me an e-mail and make us aware of the potential shortcomings of our software.
We expect this open-source software to boost research in microbial ecology based on barcoding of the ITS region, and hope that the research community will evaluate its performance also among the eukaryote groups that we have less experience with.
Metaxa in Encyclopedia of Metagenomics
A long time ago, we (Martin Eriksson, Martin Hartmann, Henrik Nilsson and me) were invited to write an overview on Metaxa for the Encyclopedia of Metagenomics. I guess the workload for pulling such a project off is huge, so there’s no surprise that it has taken a while for it to be accepted, but now it is available for consumption.
Meanwhile, Metaxa have been getting regular updates, and I hope to soon be able to show you a new major update to the software, bringing it up to the next generation of metagenomics. More on that soon.
- Bengtsson-Palme J, Hartmann M, Eriksson KM, Nilsson RH: Metaxa, overview. In:Nelson K. (Ed.) Encyclopedia of Metagenomics: SpringerReference (www.springerreference.com). Springer-Verlag Berlin Heidelberg (2013). [Link]
Server upgrades
I’ve been informed by my web service provider that there will potentially be downtime of this site on the 13th of February (Wednesday this week), due to a server upgrade. I hope this will cause as little trouble as possible (both for you and for me).
Metagenomics and Next Generation Sequencing Lab
Those attending the Metagenomics lab (part of the basic NGS course for PhD students given at GU this week), can find the material for the lab on this page:
https://microbiology.se/ngs-metagenomics-lab/
Of course, the page is open for anyone else as well, although you won’t get the support that the GU students are given.
An update to Megraft
You might remember that I a long time ago promised a minor update to Megraft. I then forgot about actually posting the update. So it’s very much about time, the updated 1.0.2 version of Megraft. The new thing in this version is improved handling of sequences with N’s (unknown bases) in them, and improved handling of sequences with strange sequence IDs (which sometimes have confused Megraft 1.0.1). The update can be downloaded here.
Metagenomics and the Hype Cycle
I was creating the diagram below an upcoming presentation, and I realized that the exponential growth in published metagenomics papers might be coming to an end. Interestingly enough the small drop in pace the recent years (701 -> 983 -> 1148) reminds me of the Hype Cycle, where we would (if my projection holds) have reached the “Peak of Inflated Expectations”, which means that we will see a rapid drop in the number of metagenomics publications in the next few years, as the field moves on.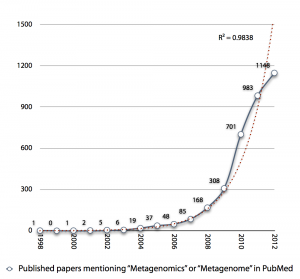
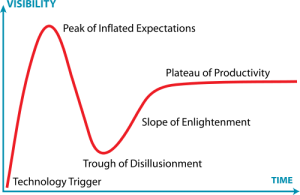 The thought is interesting, but it seems a little bit early to draw any conclusions from the number of publications, yet. It is still kind of strange to note, though, that more than 20% of metagenomics publications (740/3547) are review papers. Come on, let’s do some science first and then review it… Anyway, it’ll be interesting to see what 2013 has in store for us.
The thought is interesting, but it seems a little bit early to draw any conclusions from the number of publications, yet. It is still kind of strange to note, though, that more than 20% of metagenomics publications (740/3547) are review papers. Come on, let’s do some science first and then review it… Anyway, it’ll be interesting to see what 2013 has in store for us.
PETKit updated – Critical bug fix
Some good and some bad news regarding the PETKit. Good news first; I have written a fourth tool for the PETKit, which is included in the latest release (version 1.0.2b, download here). The new tool is called Pesort, and sorts input read pairs (or single reads) so that the read pairs occur in the same order. It also sorts out which reads that don’t have a pair and outputs them to a separate file. All this is useful if you for some reason have ended up with a scrambled read file (pair). This can e.g. happen if you want to further process the reads after running Khmer or investigate the reads remaining after mapping to a genome.
Then the bad news. There’s a critical bug in PETKit version 1.0.1b. This bug manifest itself when using custom offsets for quality scores (using the –offset option), and makes the Pearf and Pepp tools too strict – leading to that they discard reads that actually are of good quality. This does not affect the Pefcon program. If you use the PETKit for read filtering or ORF prediction, and have used custom offset values, I recommend that you re-run your data with the newly released PETKit version (1.0.2b), in which this bug has been fixed. If you have only used the default offset setting, your safe. I sincerely apologize for any inconveniences that this might have caused.