Metaxa in Encyclopedia of Metagenomics
A long time ago, we (Martin Eriksson, Martin Hartmann, Henrik Nilsson and me) were invited to write an overview on Metaxa for the Encyclopedia of Metagenomics. I guess the workload for pulling such a project off is huge, so there’s no surprise that it has taken a while for it to be accepted, but now it is available for consumption.
Meanwhile, Metaxa have been getting regular updates, and I hope to soon be able to show you a new major update to the software, bringing it up to the next generation of metagenomics. More on that soon.
- Bengtsson-Palme J, Hartmann M, Eriksson KM, Nilsson RH: Metaxa, overview. In:Nelson K. (Ed.) Encyclopedia of Metagenomics: SpringerReference (www.springerreference.com). Springer-Verlag Berlin Heidelberg (2013). [Link]
Server upgrades
I’ve been informed by my web service provider that there will potentially be downtime of this site on the 13th of February (Wednesday this week), due to a server upgrade. I hope this will cause as little trouble as possible (both for you and for me).
Metagenomics and Next Generation Sequencing Lab
Those attending the Metagenomics lab (part of the basic NGS course for PhD students given at GU this week), can find the material for the lab on this page:
https://microbiology.se/ngs-metagenomics-lab/
Of course, the page is open for anyone else as well, although you won’t get the support that the GU students are given.
An update to Megraft
You might remember that I a long time ago promised a minor update to Megraft. I then forgot about actually posting the update. So it’s very much about time, the updated 1.0.2 version of Megraft. The new thing in this version is improved handling of sequences with N’s (unknown bases) in them, and improved handling of sequences with strange sequence IDs (which sometimes have confused Megraft 1.0.1). The update can be downloaded here.
Metagenomics and the Hype Cycle
I was creating the diagram below an upcoming presentation, and I realized that the exponential growth in published metagenomics papers might be coming to an end. Interestingly enough the small drop in pace the recent years (701 -> 983 -> 1148) reminds me of the Hype Cycle, where we would (if my projection holds) have reached the “Peak of Inflated Expectations”, which means that we will see a rapid drop in the number of metagenomics publications in the next few years, as the field moves on.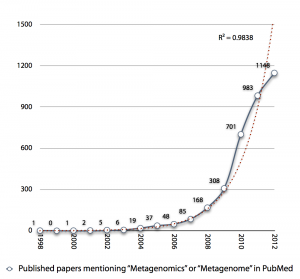
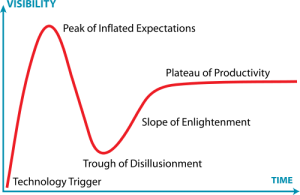 The thought is interesting, but it seems a little bit early to draw any conclusions from the number of publications, yet. It is still kind of strange to note, though, that more than 20% of metagenomics publications (740/3547) are review papers. Come on, let’s do some science first and then review it… Anyway, it’ll be interesting to see what 2013 has in store for us.
The thought is interesting, but it seems a little bit early to draw any conclusions from the number of publications, yet. It is still kind of strange to note, though, that more than 20% of metagenomics publications (740/3547) are review papers. Come on, let’s do some science first and then review it… Anyway, it’ll be interesting to see what 2013 has in store for us.
PETKit updated – Critical bug fix
Some good and some bad news regarding the PETKit. Good news first; I have written a fourth tool for the PETKit, which is included in the latest release (version 1.0.2b, download here). The new tool is called Pesort, and sorts input read pairs (or single reads) so that the read pairs occur in the same order. It also sorts out which reads that don’t have a pair and outputs them to a separate file. All this is useful if you for some reason have ended up with a scrambled read file (pair). This can e.g. happen if you want to further process the reads after running Khmer or investigate the reads remaining after mapping to a genome.
Then the bad news. There’s a critical bug in PETKit version 1.0.1b. This bug manifest itself when using custom offsets for quality scores (using the –offset option), and makes the Pearf and Pepp tools too strict – leading to that they discard reads that actually are of good quality. This does not affect the Pefcon program. If you use the PETKit for read filtering or ORF prediction, and have used custom offset values, I recommend that you re-run your data with the newly released PETKit version (1.0.2b), in which this bug has been fixed. If you have only used the default offset setting, your safe. I sincerely apologize for any inconveniences that this might have caused.
Metaxa updated to version 1.1.2
Some users have asked me to fix a table output bug in Metaxa, and I have finally got around to do so. The fix is released today in the 1.1.2 Metaxa package (download here). This version also brings an updated manual (finally), as the User’s Guide has lagged behind since version 1.0. Please continue to report bugs to metaxa [at sign] microbiology [dot] se
Bloutminer updated
Good news for everyone using my bloutminer script; it has received an update making it even more useful! Basically, I have added a function to extract the top N matches to each query (using the -n option), and I have also added the ability to output a filtered set of sequences in the same tabulated BLAST-format as the input came in. Thereby, bloutminer can now be used in more settings to easily filter out a subset in a large BLAST report (in tabular format, generated using the blastall -m 8 option). The script can be downloaded here: https://microbiology.se/software/
Introducing the PETKit
You know the feeling when your assembler supports paired-end sequences, but your FASTQ quality filterer doesn’t care about what pairs that belong together? Meaning that you end up with a mess of sequences that you have to script together in some way. Gosh, that feeling is way too common. It is for situations like that I have put together the Paired-End ToolKit (PETKit), a collection of FASTQ/FASTA sequence handling programs written in Perl. Currently the toolkit contains three command-line tools that does sequence conversion, quality filtering, and ORF prediction, all adapted for paired-end sequences specifically. You can read more about the programs, which are released as open source software, on the PETKit page. At the moment they lack proper documentation, but running the software with the “–help” option should bring up a useful set of options for each tool. This is still considered beta-software, so any bug reports, and especially suggestions, are welcome.
Also, if you have an idea of another problem that is unsolved or badly executed for paired-end sequences, let me know, and I will see if I can implement it in PETKit.
Looking for a job?
The Core Facilites at Sahlgrenska are looking for a skilled bioinformatician that can support research projects employing the Core Facilites’ services. The employee will e.g. deal with setting up analysis pipelines for next generation sequencing data. They (of course) want an experienced bioinformatician, who also knows programming (Java, C and/or C++, and scripting languages such as Perl or Python). It is also preferable if the applicant knows how to set up secure systems and manage work with the Unix/Linux terminal. More on the position can be found at GU’s web site. The application time closes on the 17th of September.