Published paper: Predicting the uncharacterized resistome
Over the weekend, Microbiome put online my most recent paper (1) – a project which started as an idea I got when I finished up my PhD thesis in 2016. One of my main points in the thesis (2), which was also made again on our recent review on environmental factors influencing resistance development (3), is that the greatest risks associated with antibiotic resistance in the environment may not be the resistance genes already circulating in pathogens (which are relatively easily quantified), but the ones associated with recruitment of novel resistance genes from bacteria in the environment (2-4). The latter genes are, however, impossible to quantify due to the fact that they are unknown. But what if we could use knowledge of the diversity and abundance of known resistance genes to estimate the same properties of the yet uncharacterized resistome? That would be a great advantage in e.g. ranking of risk environments, as then some property that is easily monitored can be used to inform risk management of both known and unknown resistance factors.
This just published paper explores this possibility, by quantifying the abundance and diversity of resistance genes in 1109 metagenomes from various environments (1). I have taken two different approaches. First, I took out smaller subsets of genes from the reference database (in this case Resqu, a database of antibiotic resistance genes with verified resistance functions, detected on mobile genetic elements), and used those subsets to estimate resistome diversity and abundance in the 1109 metagenomes. Then these predictions were compared to the results of the entire database. I then, in a second step, investigated if these predictions could be extended to a set of truly novel resistance genes, i.e. the resistance genes present in the FARME database, collecting data from functional metagenomics inserts (5,6).
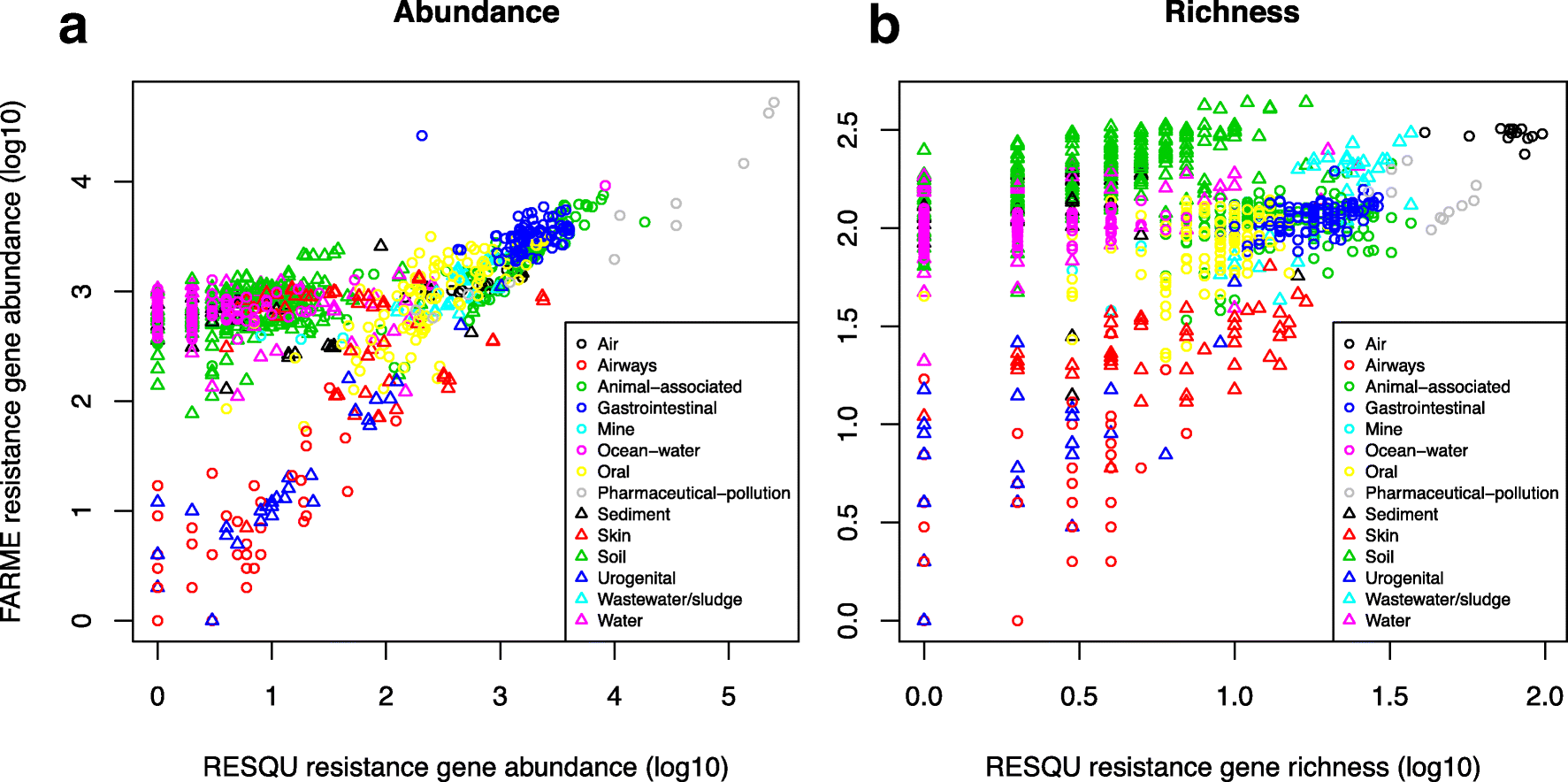
The results show that generally the diversity and abundance of known antibiotic resistance genes can be used to predict the same properties of undescribed resistance genes (see figure above). However, the extent of this predictability is, importantly, dependent on the type of environment investigated. The study also shows that carefully selected small sets of resistance genes can describe total resistance gene diversity remarkably well. This means that knowledge gained from large-scale quantifications of known resistance genes can be utilized as a proxy for unknown resistance factors. This is important for current and proposed monitoring efforts for environmental antibiotic resistance (7-11) and has implications for the design of risk ranking strategies and the choices of measures and methods for describing resistance gene abundance and diversity in the environment.
The study also investigated which diversity measures were best suited to estimate total diversity. Surprisingly, some diversity measures described the total diversity of resistance genes remarkably bad. Most prominently, the Simpson diversity index consistently showed poor performance, and while the Shannon index performed relatively better, there is still no reason to select the Shannon index over normalized (rarefied) richness of resistance genes. The ACE estimator fluctuated substantially compared to the other diversity measures, while the Chao1 estimator more consistently showed performance very similar to richness. Therefore, either richness or the Chao1 estimator should be used for ranking resistance gene diversity, while the Shannon, Simpson, and ACE measures should be avoided.
Importantly, this study implies that the recruitment of novel antibiotic resistance genes from the environment to human pathogens is essentially random. Therefore, when ranking risks associated with antibiotic resistance in environmental settings, the knowledge gained from large-scale quantification of known resistance genes can be utilized as a proxy for the unknown resistance factors (although this proxy is not perfect). Thus, high-risk environments for resistance development and dissemination would, for example, be aquaculture, animal husbandry, discharges from antibiotic manufacturing, and untreated sewage (3,8,12-15). Further attention should probably be paid to antibiotic contaminated soils, as this study points to soils as a vast source of resistance genes not yet encountered in human pathogens. This has also been suggested previously by others (16-19). The results of this study can be used to guide monitoring efforts for environmental antibiotic resistance, to design risk ranking strategies, and to choose appropriate measures and methods for describing resistance gene abundance and diversity in the environment. The entire open access paper is available here.
References
- Bengtsson-Palme J: The diversity of uncharacterized antibiotic resistance genes can be predicted from known gene variants – but not always. Microbiome, 6, 125 (2018). doi: 10.1186/s40168-018-0508-2
- Bengtsson-Palme J: Antibiotic resistance in the environment: a contribution from metagenomic studies. Doctoral thesis (medicine), Department of Infectious Diseases, Institute of Biomedicine, Sahlgrenska Academy, University of Gothenburg, 2016. [Link]
- Bengtsson-Palme J, Kristiansson E, Larsson DGJ: Environmental factors influencing the development and spread of antibiotic resistance. FEMS Microbiology Reviews, 42, 1, 68–80 (2018). doi: 10.1093/femsre/fux053
- Bengtsson-Palme J, Larsson DGJ: Antibiotic resistance genes in the environment: prioritizing risks. Nature Reviews Microbiology, 13, 369 (2015). doi: 10.1038/nrmicro3399-c1
- Wallace JC, Port JA, Smith MN, Faustian EM: FARME DB: a functional antibiotic resistance element database. Database, 2017, baw165 (2017).
- Handelsman J, Rondon MR, Brady SF, Clardy J, Goodman RM: Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chemical Biology, 5, R245–249 (1998).
- Berendonk TU, Manaia CM, Merlin C, Fatta-Kassinos D, Cytryn E, Walsh F, et al.: Tackling antibiotic resistance: the environmental framework. Nature Reviews Microbiology, 13, 310–317 (2015).
- Pruden A, Larsson DGJ, Amézquita A, Collignon P, Brandt KK, Graham DW, et al.: Management options for reducing the release of antibiotics and antibiotic resistance genes to the environment. Environmental Health Perspectives, 121, 878–885 (2013).
- Review on Antimicrobial Resistance: Antimicrobials in agriculture and the environment: reducing unnecessary use and waste. O’Neill J, ed. London: Wellcome Trust & HM Government (2015).
- Angers-Loustau A, Petrillo M, Bengtsson-Palme J, Berendonk T, Blais B, Chan KG, Coque TM, Hammer P, Heß S, Kagkli DM, Krumbiegel C, Lanza VF, Madec J-Y, Naas T, O’Grady J, Paracchini V, Rossen JWA, Ruppé E, Vamathevan J, Venturi V, Van den Eede G: The challenges of designing a benchmark strategy for bioinformatics pipelines in the identification of antimicrobial resistance determinants using next generation sequencing technologies. F1000Research, 7, 459 (2018). doi: 10.12688/f1000research.14509.1
- Larsson DGJ, Andremont A, Bengtsson-Palme J, Brandt KK, de Roda Husman AM, Fagerstedt P, Fick J, Flach C-F, Gaze WH, Kuroda M, Kvint K, Laxminarayan R, Manaia CM, Nielsen KM, Ploy M-C, Segovia C, Simonet P, Smalla K, Snape J, Topp E, van Hengel A, Verner-Jeffreys DW, Virta MPJ, Wellington EM, Wernersson A-S: Critical knowledge gaps and research needs related to the environmental dimensions of antibiotic resistance. Environment International, 117, 132–138 (2018). doi: 10.1016/j.envint.2018.04.041
- Allen HK, Donato J, Wang HH, Cloud-Hansen KA, Davies J, Handelsman J: Call of the wild: antibiotic resistance genes in natural environments. Nature Reviews Microbiology, 8, 251–259 (2010).
- Graham DW, Collignon P, Davies J, Larsson DGJ, Snape J: Underappreciated role of regionally poor water quality on globally increasing antibiotic resistance. Environmental Science & Technology, 48,11746–11747 (2014).
- Larsson DGJ: Pollution from drug manufacturing: review and perspectives. Philosophical Transactions of the Royal Society of London, Series B Biological Sciences, 369, 20130571 (2014).
- Cabello FC, Godfrey HP, Buschmann AH, Dölz HJ: Aquaculture as yet another environmental gateway to the development and globalisation of antimicrobial resistance. Lancet Infectious Diseases, 16, e127–133 (2016).
- Forsberg KJ, Reyes A, Wang B, Selleck EM, Sommer MOA, Dantas G: The shared antibiotic resistome of soil bacteria and human pathogens. Science, 337, 1107–1111 (2012).
- Allen HK, Moe LA, Rodbumrer J, Gaarder A, Handelsman J: Functional metagenomics reveals diverse beta-lactamases in a remote Alaskan soil. ISME Journal, 3, 243–251 (2009).
- Riesenfeld CS, Goodman RM, Handelsman J: Uncultured soil bacteria are a reservoir of new antibiotic resistance genes. Environmental Microbiology, 6, 981–989 (2004).
- McGarvey KM, Queitsch K, Fields S: Wide variation in antibiotic resistance proteins identified by functional metagenomic screening of a soil DNA library. Applied and Environmental Microbiology, 78, 1708–1714 (2012).
Published paper: Knowledge gaps for environmental antibiotic resistance
The outcomes from a workshop arranged by JPIAMR, the Swedish Research Council (VR) and CARe were just published as a short review paper in Environment International. In the paper, which was mostly moved forward by Prof. Joakim Larsson at CARe, we describe four major areas of knowledge gaps in the realm of environmental antibiotic resistance (1). We then highlight several important sub-questions within these areas. The broad areas we define are:
- What are the relative contributions of different sources of antibiotics and antibiotic resistant bacteria into the environment?
- What is the role of the environment as affected by anthropogenic inputs (e.g. pollution and other activities) on the evolution (mobilization, selection, transfer, persistence etc.) of antibiotic resistance?
- How significant is the exposure of humans to antibiotic resistant bacteria via different environmental routes, and what is the impact on human health?
- What technological, social, economic and behavioral interventions are effective to mitigate the emergence and spread of antibiotic resistance via the environment?
Although much has been written on the topic before (e.g. 2-12), I think it is unique that we collect and explicitly point out areas in which we are lacking important knowledge to build accurate risk models and devise appropriate intervention strategies. The workshop was held in Gothenburg on the 27–28th of September 2017. The workshop leaders Joakim Larsson, Ana-Maria de Roda Husman and Ramanan Laxminarayan were each responsible for moderating a breakout group, and every breakout group was tasked to deal with knowledge gaps related to either evolution, transmission or interventions. The reports of the breakout groups were then discussed among all participants to clarify and structure the areas where more research is needed, which boiled down to the four overarching critical knowledge gaps described in the paper (1).
This is a short paper, and I think everyone with an interest in environmental antibiotic resistance should read it and reflect over its content (because, we may of course have overlooked some important aspect). You can find the paper here.
References
- Larsson DGJ, Andremont A, Bengtsson-Palme J, Brandt KK, de Roda Husman AM, Fagerstedt P, Fick J, Flach C-F, Gaze WH, Kuroda M, Kvint K, Laxminarayan R, Manaia CM, Nielsen KM, Ploy M-C, Segovia C, Simonet P, Smalla K, Snape J, Topp E, van Hengel A, Verner-Jeffreys DW, Virta MPJ, Wellington EM, Wernersson A-S: Critical knowledge gaps and research needs related to the environmental dimensions of antibiotic resistance. Environment International, 117, 132–138 (2018). doi: 10.1016/j.envint.2018.04.041
- Bengtsson-Palme J, Kristiansson E, Larsson DGJ: Environmental factors influencing the development and spread of antibiotic resistance. FEMS Microbiology Reviews, 42, 1, 68–80 (2018). doi: 10.1093/femsre/fux053
- Martinez JL, Coque TM, Baquero F: What is a resistance gene? Ranking risk in resistomes. Nature Reviews Microbiology 2015, 13:116–123. doi:10.1038/nrmicro3399
- Bengtsson-Palme J, Larsson DGJ: Antibiotic resistance genes in the environment: prioritizing risks. Nature Reviews Microbiology, 13, 369 (2015). doi: 10.1038/nrmicro3399-c1
- Ashbolt NJ, Amézquita A, Backhaus T, Borriello P, Brandt KK, Collignon P, et al.: Human Health Risk Assessment (HHRA) for Environmental Development and Transfer of Antibiotic Resistance. Environmental Health Perspectives, 121, 993–1001 (2013)
- Pruden A, Larsson DGJ, Amézquita A, Collignon P, Brandt KK, Graham DW, et al.: Management options for reducing the release of antibiotics and antibiotic resistance genes to the environment. Environmental Health Perspectives, 121, 878–85 (2013).
- Gillings MR: Evolutionary consequences of antibiotic use for the resistome, mobilome and microbial pangenome. Frontiers in Microbiology, 4, 4 (2013).
- Baquero F, Alvarez-Ortega C, Martinez JL: Ecology and evolution of antibiotic resistance. Environmental Microbiology Reports, 1, 469–476 (2009).
- Baquero F, Tedim AP, Coque TM: Antibiotic resistance shaping multi-level population biology of bacteria. Frontiers in Microbiology, 4, 15 (2013).
- Berendonk TU, Manaia CM, Merlin C et al.: Tackling antibiotic resistance: the environmental framework. Nature Reviews Microbiology, 13, 310–317 (2015).
- Hiltunen T, Virta M, Laine A-L: Antibiotic resistance in the wild: an eco-evolutionary perspective. Philosophical Transactions of the Royal Society B: Biological Sciences, 372 (2017) doi: 10.1098/rstb.2016.0039.
- Martinez JL: Bottlenecks in the transferability of antibiotic resistance from natural ecosystems to human bacterial pathogens. Frontiers in Microbiology, 2, 265 (2011).
Published paper: A novel Na-binding site in sialic acid symporters
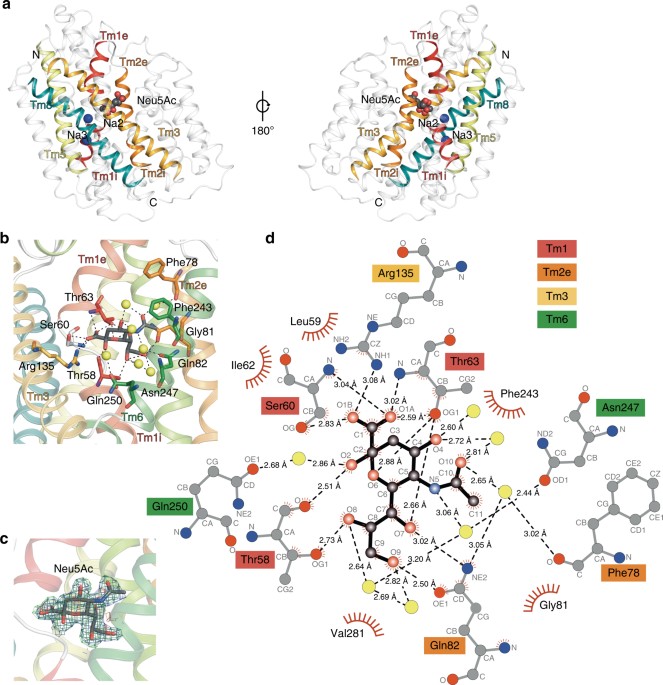
I have been quite occupied with other things the last couple of days, so I am late on the ball here. Anyway, on May 1st, Nature Communications published a paper on the protein structure of SiaT, a sialic acid transporter from Proteus mirabilis (1). Many pathogens use sialic acids as an energy source or as an external coating to evade the immune defense (2). Therefore, many bacteria that colonize sialylated environments have transporters which specifically import sialic acids. SiaT is one of those transporters, belonging to the sodium solute symporter (SSS) family (3) (with for some weird reason is associated with the Pfam family “SSF”, an eternal source of confusion in discussions within this project). The SSS proteins use Na+ gradients to drive the import of desired substrates (4). Based on the protein structure, our team found that SiaT binds two Na+ ions. One binds to the conserved, well-known, Na2 site, but the other Na+ binds to a new position, which we term Na3. This position (this is where my part of the work comes in) is conserved in many SSS family members. We finally used functional and molecular dynamics studies to validate the substrate-binding site and demonstrate that both Na+ sites regulate N-acetylneuraminic acid transport.
As I hinted, i am not venturing into protein structures – that part of this work has been performed by an excellent team associated with Dr. Rosmarie Friemann. Instead, my part is essentially summarized in these two sentences of the manuscript: “We analysed all SSS sequences that contained the primary Na2 site (21,467) to determine the degree of conservation of the Na3 site, allowing for threonine at either Ser345 or Ser346. Na3 is present in 19.6% (4212) of these sequences including hSGLT1, which transports two Na+, but not vSGLT or hSGLT2, which transport only one Na+” (1). That’s a few months of works condensed into 55 words. Still, the exciting thing here is that we find an evolutionary conserved Na-binding site, which has so far eluded detection.
The results of this work provides a better understanding of how secondary active transporters harness additional energy from ion gradients. It may be possible to exploit differences in this mechanism between different SSS family members (and other transporters with the LeuT fold) to develop new antimicrobials, something that is urgently needed in the face of the rapidly increasing antibiotic resistance.
References
- Wahlgren WY°, North RA°, Dunevall E°, Paz A, Scalise M, Bisognano P, Bengtsson-Palme J, Goyal P, Claesson E, Caing-Carlsson R, Andersson R, Beis K, Nilsson U, Farewell A, Pochini L, Indiveri C, Grabe M, Dobson RCJ, Abramson J, Ramaswamy S, Friemann R: Substrate-bound outward-open structure of a Na+-coupled sialic acid symporter reveals a novel Na+ site. Nature Communications, 9, 1753 (2018). doi: 10.1038/s41467-018-04045-7
- Vimr ER, Kalivoda KA, Deszo EL, Steenburgen SM: Diversity of microbial sialic acid metabolism. Microbiology and Molecular Biology Reviews, 68, 132–153 (2004).
- North RA, Horne CR, Davies JS, Remus DM, Muscroft-Taylor AC, Goyal P, Wahlgren WY, Ramaswamy S, Friemann R, Dobson RCJ: “Just a spoonful of sugar…”: import of sialic acid across bacterial cell membranes. Biophysical Reviews, 10, 219–227 (2017).
- Severi E, Hosie AH, Hawkhead JA, Thomas GH: Characterization of a novel sialic acid transporter of the sodium solute symporter (SSS) family and in vivo comparison with known bacterial sialic acid transporters. FEMS Microbiology Letters, 304, 47–54 (2010).
Published paper: Selective concentrations for ciprofloxacin
My colleagues in Gothenburg have published a new paper in Environment International, in which I was involved in the bioinformatics analyses. In the paper, for which Nadine Kraupner did the lion’s share of the work, we establish minimal selective concentrations (MSCs) for resistance to the antibiotic ciprofloxacin in Escherichia coli grown in complex microbial communities (1). We also determine the community responses at the taxonomic and resistance gene levels. Nadine has made use of Sara Lundström’s aquarium system (2) to grow biofilms in the exposure of sublethal levels of antibiotics. Using the system, we find that 1 μg/L ciprofloxacin selects for the resistance gene qnrD, while 10 μg/L ciprofloxacin is required to detect changes of phenotypic resistance. In short, the different endpoints studied (and their corresponding MSCs) were:
- CFU counts from test tubes, grown on R2A plates with 2 mg/L ciprofloxain – MSC = 5 μg/L
- CFU counts from aquaria, grown on R2A plates with 0.25 or 2 mg/L ciprofloxain – MSC = 10 μg/L
- Chromosomal resistance mutations – MSC ~ 10 μg/L
- Increased resistance gene abundances, metagenomics – MSC range: 1 μg/L
- Changes to taxonomic diversity – 1 µg/L
- Changes to taxonomic community composition – MSC ~ 1-10 μg/L
We have previously reported a predicted no-effect concentration for resistance of 0.064 µg/L for ciprofloxacin (3), which corresponds fairly well with the MSCs determined experimentally here, being around a factor of ten off. However, we cannot exclude that in other experimental systems, the selective effects of ciprofloxacin could be even lower and thus the predicted PNEC may still be relevant. The selective concentrations we report for ciprofloxacin are close to those that have been reported in sewage treatment plants (3-5), suggesting the possibility for weak selection of resistance. Several recent reports have underscored the need to populate the this far conceptual models for resistance development in the environment with actual numbers (6-10). Determining selective concentrations for different antibiotics in actual community settings is an important step on the road towards building accurate quantitative models for resistance emergence and propagation.
References
- Kraupner N, Ebmeyer S, Bengtsson-Palme J, Fick J, Kristiansson E, Flach C-F, Larsson DGJ: Selective concentration for ciprofloxacin in Escherichia coli grown in complex aquatic bacterial biofilms. Environment International, 116, 255–268 (2018). doi: 10.1016/j.envint.2018.04.029 [Paper link]
- Lundström SV, Östman M, Bengtsson-Palme J, Rutgersson C, Thoudal M, Sircar T, Blanck H, Eriksson KM, Tysklind M, Flach C-F, Larsson DGJ: Minimal selective concentrations of tetracycline in complex aquatic bacterial biofilms. Science of the Total Environment, 553, 587–595 (2016). doi: 10.1016/j.scitotenv.2016.02.103 [Paper link]
- Bengtsson-Palme J, Larsson DGJ: Concentrations of antibiotics predicted to select for resistant bacteria: Proposed limits for environmental regulation. Environment International, 86, 140-149 (2016). doi: 10.1016/j.envint.2015.10.015
- Michael I, Rizzo L, McArdell CS, Manaia CM, Merlin C, Schwartz T, Dagot C, Fatta-Kassinos D: Urban wastewater treatment plants as hotspots for the release of antibiotics in the environment: a review. Water Research, 47, 957–995 (2013). doi:10.1016/j.watres.2012.11.027
- Bengtsson-Palme J, Hammarén R, Pal C, Östman M, Björlenius B, Flach C-F, Kristiansson E, Fick J, Tysklind M, Larsson DGJ: Elucidating selection processes for antibiotic resistance in sewage treatment plants using metagenomics. Science of the Total Environment, 572, 697–712 (2016). doi: 10.1016/j.scitotenv.2016.06.228
- Ågerstrand M, Berg C, Björlenius B, Breitholtz M, Brunstrom B, Fick J, Gunnarsson L, Larsson DGJ, Sumpter JP, Tysklind M, Rudén C: Improving environmental risk assessment of human pharmaceuticals. Environmental Science and Technology (2015). doi:10.1021/acs.est.5b00302
- Bengtsson-Palme J, Kristiansson E, Larsson DGJ: Environmental factors influencing the development and spread of antibiotic resistance. FEMS Microbiology Reviews, 42, 1, 68–80 (2018). doi: 10.1093/femsre/fux053
- Joint Programming Initiative on Antimicrobial Resistance: JPIAMR Workshop on Environmental Dimensions of AMR: Summary and recommendations. JPIAMR (2017). [Link]
- Angers A, Petrillo P, Patak, A, Querci M, Van den Eede G: The Role and Implementation of Next-Generation Sequencing Technologies in the Coordinated Action Plan against Antimicrobial Resistance. JRC Conference and Workshop Report, EUR 28619 (2017). doi: 10.2760/745099
- Larsson DGJ, Andremont A, Bengtsson-Palme J, Brandt KK, de Roda Husman AM, Fagerstedt P, Fick J, Flach C-F, Gaze WH, Kuroda M, Kvint K, Laxminarayan R, Manaia CM, Nielsen KM, Ploy M-C, Segovia C, Simonet P, Smalla K, Snape J, Topp E, van Hengel A, Verner-Jeffreys DW, Virta MPJ, Wellington EM, Wernersson A-S: Critical knowledge gaps and research needs related to the environmental dimensions of antibiotic resistance. Environment International, in press (2018). doi: 10.1016/j.envint.2018.04.041
And the experiments have started!
It’s been a long time before I have written something here, mostly because making ourselves at home in Madison have take some time; then we go the flue; and then there have been a lot at work after that. But now i will try to have another go at writing somethings on the Wisconsin Blog. First, a look at my (already messy) desk here at the Wisconsin Institute for Discovery.
And then, a look at my lab space, which I have a view of straight from my desk, through a glass window.
This week, I have started experiments with exposing our little model community to antibiotics and it looks like I’m getting potentially exciting results. I have to sit down with the data today to see if there’s statistical differences, but from the looks of the biofilms, there is potential here.
Next week I will try to start experiment with sand columns and see if I can replicate some of this in this setting as well. It is interesting being back in the lab, and I feel that this an experience that will be very valuable for me going forward. I look forward to the days later this spring when I will start generating sequence data from my own experiments!
Published opinion piece: Protection goals and risk assessment
Recently, Le Page et al. published a paper in Environmental International (1), partially building on the predicted no-effect concentrations for resistance selection for 111 antibiotics that me and Joakim Larsson published around two years ago (2). In their paper, the authors stress that discharge limits for antibiotics need to consider their potency to affect both environmental and human health, which we believe is a very reasonable standpoint, and to which we agree. However, we do not agree on the authors’ claim that cyanobacteria would often be more sensitive to antibiotics than the most sensitive human-associated bacteria (1). Importantly, we also think that it is a bit unclear from the paper which protection goals are considered. Are the authors mainly concerned with protecting microbial diversity in ecosystems, protecting ecosystem functions and services, or protecting from risks for resistance selection? This is important because it influence why one would want to mitigate, and therefore who would perform which actions. To elaborate a little on our standpoints, we wrote a short correspondence piece to Environment International, which is now published (3). (It has been online for a few days, but without a few last-minute changes we did to the proof, and hence I’m only posting about it now when the final version is online.) There is indeed an urgent need for discharge limits for antibiotics, particularly for industrial sources (4) and such limits would have tremendous value in regulation efforts, and in development of environmental criteria within public procurement and generic exchange programs (5). Importantly, while we are all for taking ecotoxicological data into account when doing risk assessment, we think that there should be solid scientific ground for mitigations and that regulations need to consider the benefits versus the costs, which is what we want to convey in our response to Le Page et al.
References
- Le Page G, Gunnarsson L, Snape J, Tyler CR: Integrating human and environmental health in antibiotic risk assessment: a critical analysis of protection goals, species sensitivity and antimicrobial resistance. Environment International, in press (2017). doi: 10.1016/j.envint.2017.09.013
- Bengtsson-Palme J, Larsson DGJ: Concentrations of antibiotics predicted to select for resistant bacteria: Proposed limits for environmental regulation. Environment International, 86, 140–149 (2016). doi: 10.1016/j.envint.2015.10.015
- Bengtsson-Palme J, Larsson DGJ: Protection goals must guide risk assessment for antibiotics. Environment International, in press (2017). doi: 10.1016/j.envint.2017.10.019
- Bengtsson-Palme J, Larsson DGJ: Time to limit antibiotic pollution. The Medicine Maker, 0416, 302, 17–18 (2016). [Paper link]
- Bengtsson-Palme J, Gunnarsson L, Larsson DGJ: Can branding and price of pharmaceuticals guide informed choices towards improved pollution control during manufacturing? Journal of Cleaner Production, 171, 137–146 (2018). doi: 10.1016/j.jclepro.2017.09.247
Published paper: Drug price is linked to environmental standards
Yesterday, Swedish television channel TV4 highlighted a recent publication by myself, Lina Gunnarsson and Joakim Larsson, in which we show that the price of pharmaceuticals is linked to the environmental standards of production countries. Surprisingly, however, this link seems to be mostly driven by whether the product is generic or original (branded), which in turns affect the prices.
In the study (1), published in Journal of Cleaner Production, we have used an exclusive set of Swedish sales data for pharmaceuticals combined with data on the origin of the active ingredients, obtained under an agreement to not identify individual manufacturers or products. We used this data to determine if price pressure and generic substitution could be linked to the general environmental performance and the corruption levels of the production countries, as measured by the Environmental Performance Index (2) and the Corruption Perception Index (3). In line with what we believed, India was the largest producer of generics, while Europe and the USA dominated the market for branded products (1). Importantly, we found that the price and environmental performance index of the production countries were linked, but that this relationship was largely explained by whether the product was original or generic.
To some extent, this relationship would allow buyers to select products that likely originate from countries that, in general terms, have better pollution control, which was also highlighted in the news clip that TV4 produced. However, what was lacking from that clip was the fact that this approach lacks resolution, because it does not say anything about the environmental footprint of individual products. We therefore conclude that to better allow consumers, hospitals and pharmacies to influence the environmental impact of their product choices, there is need for regulation and, importantly, transparency in the production chain, as has also been pointed out earlier (4,5). To this end, emissions from manufacturing need to be measured, allowing for control and follow-up on industry commitments towards sustainable manufacturing of pharmaceuticals (6). Since the discharges from pharmaceutical manufacturing not only leads to consequences to the local environment (7,8), but also in the case of antibiotics has potentially global consequences in terms of increasing risks for resistance development (9), limiting discharges is an urgent need to avoid a looming antibiotic resistance crisis (10).
The paper was also highlighted by the Centre for Antibiotic Resistance Research, and can be read here or here.
References
- Bengtsson-Palme J, Gunnarsson L, Larsson DGJ: Can branding and price of pharmaceuticals guide informed choices towards improved pollution control during manufacturing? Journal of Cleaner Production, 171, 137–146 (2018). doi: 10.1016/j.jclepro.2017.09.247
- Hsu A, Alexandre N, Cohen S, Jao P, Khusainova E: 2016 Environmental Performance Index. Yale University, New Haven, CT, USA (2016). http://epi.yale.edu/reports/2016-report
- Transparency International: Corruption Perceptions Index 2014. Transparency International, Berlin, Germany (2014). http://www.transparency.org/cpi2014/in_detail
- Larsson DGJ, Fick J: Transparency throughout the production chain–a way to reduce pollution from the manufacturing of pharmaceuticals? Regulatory Toxicology and Pharmacology, 53, 161–163 (2009). doi:10.1016/j.yrtph.2009.01.008
- Ågerstrand M, Berg C, Björlenius B, Breitholtz M, Brunström B, Fick J, Gunnarsson L, Larsson DGJ, Sumpter JP, Tysklind M, Rudén C: Improving environmental risk assessment of human pharmaceuticals. Environmental Science & Technology, 49, 5336–5345 (2015). doi:10.1021/acs.est.5b00302
- Industry Roadmap for Progress on Combating Antimicrobial Resistance: Industry Roadmap for Progress on Combating Antimicrobial Resistance – September 2016. (2016). http://www.ifpma.org/wp-content/uploads/2016/09/Roadmap-for-Progress-on-AMR-FINAL.pdf
- Larsson DGJ, de Pedro C, Paxeus N: Effluent from drug manufactures contains extremely high levels of pharmaceuticals. Journal of Hazardous Materials, 148, 751–755 (2007). doi:10.1016/j.jhazmat.2007.07.008
- aus der Beek T, Weber FA, Bergmann A, Hickmann S, Ebert I, Hein A, Küster A: Pharmaceuticals in the environment–Global occurrences and perspectives. Environmental Toxicology and Chemistry, 35, 823–835 (2016). doi:10.1002/etc.3339
- Bengtsson-Palme J, Larsson DGJ: Concentrations of antibiotics predicted to select for resistant bacteria: Proposed limits for environmental regulation. Environment International, 86, 140–149 (2016). doi: 10.1016/j.envint.2015.10.015
- Bengtsson-Palme J, Larsson DGJ: Time to limit antibiotic pollution. The Medicine Maker, 0416, 302, 17–18 (2016). [Paper link]
Published paper: Environmental factors leading to resistance
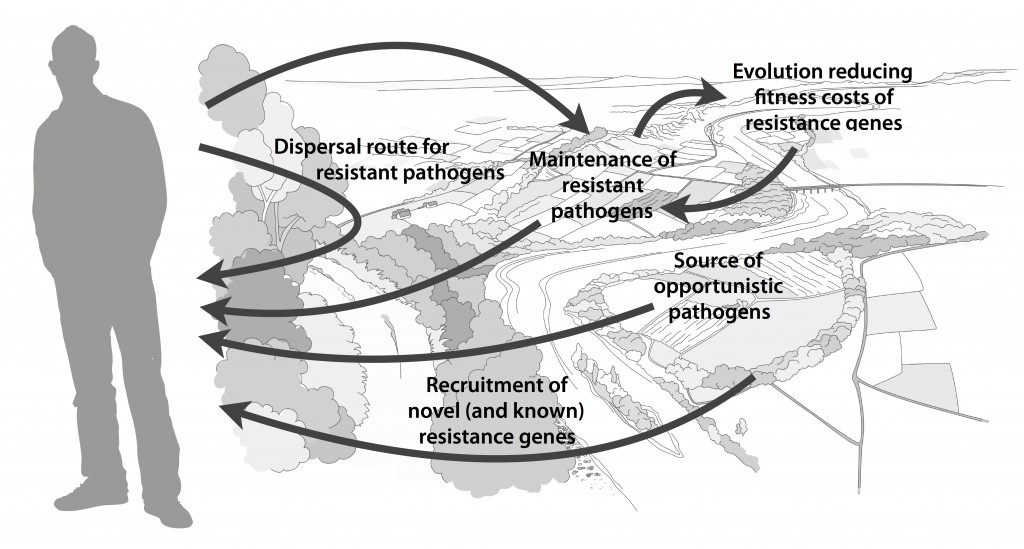
Myself, Joakim Larsson and Erik Kristiansson have written a review on the environmental factors that influence development and spread of antibiotic resistance, which was published today in FEMS Microbiology Reviews. The review (1) builds on thoughts developed in the latter parts of my PhD thesis (2), and seeks to provide a synthesis knowledge gained from different subfields towards the current understanding of evolutionary and ecological processes leading to clinical appearance of resistance genes, as well as the important environmental dispersal barriers preventing spread of resistant pathogens.
We postulate that emergence of novel resistance factors and mobilization of resistance genes are likely to occur continuously in the environment. However, the great majority of such genetic events are unlikely to lead to establishment of novel resistance factors in bacterial populations, unless there is a selection pressure for maintaining them or their fitness costs are negligible. To enable measures to prevent resistance development in the environment, it is therefore critical to investigate under what conditions and to what extent environmental selection for resistance takes place. Selection for resistance is likely less important for the dissemination of resistant bacteria, but will ultimately depend on how well the species or strain in question thrives in the external environment. Metacommunity theory (3,4) suggests that dispersal ability is central to this process, and therefore opportunistic pathogens with their main habitat in the environment may play an important role in the exchange of resistance factors between humans and the environment. Understanding the dispersal barriers hindering this exchange is not only key to evaluate risks, but also to prevent resistant pathogens, as well as novel resistance genes, from reaching humans.
Towards the end of the paper, we suggest certain environments that seem to be more important from a risk management perspective. We also discuss additional problems linked to the development of antibiotic resistance, such as increased evolvability of bacterial genomes (5) and which other types of genes that may be mobilized in the future, should the development continue (1,6). In this review, we also further develop thoughts on the relative risks of re-recruiting and spreading well-known resistance factors already circulating in pathogens, versus recruitment of completely novel resistance genes from environmental bacteria (7). While the latter case is likely to be very rare, and thus almost impossible to quantify the risks for, the consequences of such (potentially one-time) events can be dire.
I personally think that this is one of the best though-through pieces I have ever written, and since it is open access and (in my biased opinion) written in a fairly accessible way, I recommend everyone to read it. It builds on the ecological theories for resistance ecology developed by, among others, Fernando Baquero and José Martinez (8-13). Over the last year, it has been stressed several times at meetings (e.g. at the EDAR conferences in August) that there is a need to develop an ecological framework for antibiotic resistance genes. I think this paper could be one of the foundational pillars on such an endeavor and look forward to see how it will fit into the growing literature on the subject!
References
- Bengtsson-Palme J, Kristiansson E, Larsson DGJ: Environmental factors influencing the development and spread of antibiotic resistance. FEMS Microbiology Reviews, accepted manuscript (2017). doi: 10.1093/femsre/fux053
- Bengtsson-Palme J: Antibiotic resistance in the environment: a contribution from metagenomic studies. Doctoral thesis (medicine), Department of Infectious Diseases, Institute of Biomedicine, Sahlgrenska Academy, University of Gothenburg, 2016. [Link]
- Bengtsson J: Applied (meta)community ecology: diversity and ecosystem services at the intersection of local and regional processes. In: Verhoef HA, Morin PJ (eds.). Community Ecology: Processes, Models, and Applications. Oxford: Oxford University Press, 115–130 (2009).
- Leibold M, Norberg J: Biodiversity in metacommunities: Plankton as complex adaptive systems? Limnology and Oceanography, 1278–1289 (2004).
- Gillings MR, Stokes HW: Are humans increasing bacterial evolvability? Trends in Ecology and Evolution, 27, 346–352 (2012).
- Gillings MR: Evolutionary consequences of antibiotic use for the resistome, mobilome and microbial pangenome. Frontiers in Microbiology, 4, 4 (2013).
- Bengtsson-Palme J, Larsson DGJ: Antibiotic resistance genes in the environment: prioritizing risks. Nature Reviews Microbiology, 13, 369 (2015). doi: 10.1038/nrmicro3399-c1
- Baquero F, Alvarez-Ortega C, Martinez JL: Ecology and evolution of antibiotic resistance. Environmental Microbiology Reports, 1, 469–476 (2009).
- Baquero F, Tedim AP, Coque TM: Antibiotic resistance shaping multi-level population biology of bacteria. Frontiers in Microbiology, 4, 15 (2013).
- Berendonk TU, Manaia CM, Merlin C et al.: Tackling antibiotic resistance: the environmental framework. Nature Reviews Microbiology, 13, 310–317 (2015).
- Hiltunen T, Virta M, Laine A-L: Antibiotic resistance in the wild: an eco-evolutionary perspective. Philosophical Transactions of the Royal Society B: Biological Sciences, 372 (2017) doi: 10.1098/rstb.2016.0039.
- Martinez JL: Bottlenecks in the transferability of antibiotic resistance from natural ecosystems to human bacterial pathogens. Frontiers in Microbiology, 2, 265 (2011).
- Salyers AA, Amábile-Cuevas CF: Why are antibiotic resistance genes so resistant to elimination? Antimicrobial Agents and Chemotherapy, 41, 2321–2325 (1997).
Talk on emission limits in Stockholm
In two weeks time, on the 15th of June, I will participate in a seminar organised by Landstingens nätverk för läkemedel och miljö (the Swedish county council network for pharmaceuticals and environment; the seminar will be held in Swedish) in Stockholm. I will give a talk on our proposed emission limits for antibiotics published last year (the paper is available here), but there will also be talks on wastewater treatment, sustainable pharmaceutical usage and environmental standards for pharmaceuticals. The full program can be found here, and you may register here until June 9. The seminar is free of charge.
And if you are interested in this, I can also recommend the webinar given by Healthcare Without Harm next week (on June 8), which will deal with sustainable procurement as a means to deal with pharmaceutical pollution in the environment. I will at least tune in to hear how the discussion goes here.
Webinar online and the Science Festival
First of all, I am happy to announce that the webinar I participated in on the (un)recognised pathways of AMR: Air pollution and food, organised by Healthcare Without Harm is now put online so that you can view it, in case you missed out on this event. To be honest it is probably not one of my best public appearances, but the topic is highly interesting.
Second, next week I am taking part in Vetenskapsfestivalen – the Science Festival in Gothenburg. Specifically, I will be on of the researchers participating in the Science Roulette, taking place in the big ferris wheel at Liseberg. This will take place between 17.00 and 18.00 on May 11th. The idea is that people will be paired with researchers in diverse subjects, of which I am one, and then have a 20 minute chat while the wheel is spinning. Sounds like potential for lot of fun, and I hope to see you there! I will discuss antibiotic resistance, and for how much longer we can trust that our antibiotics will work.