Published paper: Annotating fungi from the built environment part II
MycoKeys earlier this week published a paper describing the results of a workshop in Aberdeen in April last year, where we refined annotations for fungal ITS sequences from the built environment (1). This was a follow-up on a workshop in May 2016 (2) and the results have been implemented in the UNITE database and shared with other online resources. The paper has also been highlighted at microBEnet. I have very little time to further comment on this at this very moment, but I believe, as I wrote last time, that distributed initiatives like this (and the ones I have been involved in in the past (3,4)) serve a very important purpose for establishing better annotation of sequence data (5). The full paper can be found here.
References
- Nilsson RH, Taylor AFS, Adams RI, Baschien C, Bengtsson-Palme J, Cangren P, Coleine C, Daniel H-M, Glassman SI, Hirooka Y, Irinyi L, Iršenaite R, Martin-Sánchez PM, Meyer W, Oh S-O, Sampaio JP, Seifert KA, Sklenár F, Stubbe D, Suh S-O, Summerbell R, Svantesson S, Unterseher M, Visagie CM, Weiss M, Woudenberg J, Wurzbacher C, Van den Wyngaert S, Yilmaz N, Yurkov A, Kõljalg U, Abarenkov K: Annotating public fungal ITS sequences from the built environment according to the MIxS-Built Environment standard – a report from an April 10-11, 2017 workshop (Aberdeen, UK). MycoKeys, 28, 65–82 (2018). doi: 10.3897/mycokeys.28.20887 [Paper link]
- Abarenkov K, Adams RI, Laszlo I, Agan A, Ambrioso E, Antonelli A, Bahram M, Bengtsson-Palme J, Bok G, Cangren P, Coimbra V, Coleine C, Gustafsson C, He J, Hofmann T, Kristiansson E, Larsson E, Larsson T, Liu Y, Martinsson S, Meyer W, Panova M, Pombubpa N, Ritter C, Ryberg M, Svantesson S, Scharn R, Svensson O, Töpel M, Untersehrer M, Visagie C, Wurzbacher C, Taylor AFS, Kõljalg U, Schriml L, Nilsson RH: Annotating public fungal ITS sequences from the built environment according to the MIxS-Built Environment standard – a report from a May 23-24, 2016 workshop (Gothenburg, Sweden). MycoKeys, 16, 1–15 (2016). doi: 10.3897/mycokeys.16.10000
- Kõljalg U, Nilsson RH, Abarenkov K, Tedersoo L, Taylor AFS, Bahram M, Bates ST, Bruns TT, Bengtsson-Palme J, Callaghan TM, Douglas B, Drenkhan T, Eberhardt U, Dueñas M, Grebenc T, Griffith GW, Hartmann M, Kirk PM, Kohout P, Larsson E, Lindahl BD, Lücking R, Martín MP, Matheny PB, Nguyen NH, Niskanen T, Oja J, Peay KG, Peintner U, Peterson M, Põldmaa K, Saag L, Saar I, Schüßler A, Senés C, Smith ME, Suija A, Taylor DE, Telleria MT, Weiß M, Larsson KH: Towards a unified paradigm for sequence-based identification of Fungi. Molecular Ecology, 22, 21, 5271–5277 (2013). doi: 10.1111/mec.12481
- Nilsson RH, Hyde KD, Pawlowska J, Ryberg M, Tedersoo L, Aas AB, Alias SA, Alves A, Anderson CL, Antonelli A, Arnold AE, Bahnmann B, Bahram M, Bengtsson-Palme J, Berlin A, Branco S, Chomnunti P, Dissanayake A, Drenkhan R, Friberg H, Frøslev TG, Halwachs B, Hartmann M, Henricot B, Jayawardena R, Jumpponen A, Kauserud H, Koskela S, Kulik T, Liimatainen K, Lindahl B, Lindner D, Liu J-K, Maharachchikumbura S, Manamgoda D, Martinsson S, Neves MA, Niskanen T, Nylinder S, Pereira OL, Pinho DB, Porter TM, Queloz V, Riit T, Sanchez-García M, de Sousa F, Stefaczyk E, Tadych M, Takamatsu S, Tian Q, Udayanga D, Unterseher M, Wang Z, Wikee S, Yan J, Larsson E, Larsson K-H, Kõljalg U, Abarenkov K: Improving ITS sequence data for identification of plant pathogenic fungi. Fungal Diversity, 67, 1, 11–19 (2014). doi: 10.1007/s13225-014-0291-8
- Bengtsson-Palme J, Boulund F, Edström R, Feizi A, Johnning A, Jonsson VA, Karlsson FH, Pal C, Pereira MB, Rehammar A, Sánchez J, Sanli K, Thorell K: Strategies to improve usability and preserve accuracy in biological sequence databases. Proteomics, Early view (2016). doi: 10.1002/pmic.201600034
Published opinion piece: Protection goals and risk assessment
Recently, Le Page et al. published a paper in Environmental International (1), partially building on the predicted no-effect concentrations for resistance selection for 111 antibiotics that me and Joakim Larsson published around two years ago (2). In their paper, the authors stress that discharge limits for antibiotics need to consider their potency to affect both environmental and human health, which we believe is a very reasonable standpoint, and to which we agree. However, we do not agree on the authors’ claim that cyanobacteria would often be more sensitive to antibiotics than the most sensitive human-associated bacteria (1). Importantly, we also think that it is a bit unclear from the paper which protection goals are considered. Are the authors mainly concerned with protecting microbial diversity in ecosystems, protecting ecosystem functions and services, or protecting from risks for resistance selection? This is important because it influence why one would want to mitigate, and therefore who would perform which actions. To elaborate a little on our standpoints, we wrote a short correspondence piece to Environment International, which is now published (3). (It has been online for a few days, but without a few last-minute changes we did to the proof, and hence I’m only posting about it now when the final version is online.) There is indeed an urgent need for discharge limits for antibiotics, particularly for industrial sources (4) and such limits would have tremendous value in regulation efforts, and in development of environmental criteria within public procurement and generic exchange programs (5). Importantly, while we are all for taking ecotoxicological data into account when doing risk assessment, we think that there should be solid scientific ground for mitigations and that regulations need to consider the benefits versus the costs, which is what we want to convey in our response to Le Page et al.
References
- Le Page G, Gunnarsson L, Snape J, Tyler CR: Integrating human and environmental health in antibiotic risk assessment: a critical analysis of protection goals, species sensitivity and antimicrobial resistance. Environment International, in press (2017). doi: 10.1016/j.envint.2017.09.013
- Bengtsson-Palme J, Larsson DGJ: Concentrations of antibiotics predicted to select for resistant bacteria: Proposed limits for environmental regulation. Environment International, 86, 140–149 (2016). doi: 10.1016/j.envint.2015.10.015
- Bengtsson-Palme J, Larsson DGJ: Protection goals must guide risk assessment for antibiotics. Environment International, in press (2017). doi: 10.1016/j.envint.2017.10.019
- Bengtsson-Palme J, Larsson DGJ: Time to limit antibiotic pollution. The Medicine Maker, 0416, 302, 17–18 (2016). [Paper link]
- Bengtsson-Palme J, Gunnarsson L, Larsson DGJ: Can branding and price of pharmaceuticals guide informed choices towards improved pollution control during manufacturing? Journal of Cleaner Production, 171, 137–146 (2018). doi: 10.1016/j.jclepro.2017.09.247
Published paper: Drug price is linked to environmental standards
Yesterday, Swedish television channel TV4 highlighted a recent publication by myself, Lina Gunnarsson and Joakim Larsson, in which we show that the price of pharmaceuticals is linked to the environmental standards of production countries. Surprisingly, however, this link seems to be mostly driven by whether the product is generic or original (branded), which in turns affect the prices.
In the study (1), published in Journal of Cleaner Production, we have used an exclusive set of Swedish sales data for pharmaceuticals combined with data on the origin of the active ingredients, obtained under an agreement to not identify individual manufacturers or products. We used this data to determine if price pressure and generic substitution could be linked to the general environmental performance and the corruption levels of the production countries, as measured by the Environmental Performance Index (2) and the Corruption Perception Index (3). In line with what we believed, India was the largest producer of generics, while Europe and the USA dominated the market for branded products (1). Importantly, we found that the price and environmental performance index of the production countries were linked, but that this relationship was largely explained by whether the product was original or generic.
To some extent, this relationship would allow buyers to select products that likely originate from countries that, in general terms, have better pollution control, which was also highlighted in the news clip that TV4 produced. However, what was lacking from that clip was the fact that this approach lacks resolution, because it does not say anything about the environmental footprint of individual products. We therefore conclude that to better allow consumers, hospitals and pharmacies to influence the environmental impact of their product choices, there is need for regulation and, importantly, transparency in the production chain, as has also been pointed out earlier (4,5). To this end, emissions from manufacturing need to be measured, allowing for control and follow-up on industry commitments towards sustainable manufacturing of pharmaceuticals (6). Since the discharges from pharmaceutical manufacturing not only leads to consequences to the local environment (7,8), but also in the case of antibiotics has potentially global consequences in terms of increasing risks for resistance development (9), limiting discharges is an urgent need to avoid a looming antibiotic resistance crisis (10).
The paper was also highlighted by the Centre for Antibiotic Resistance Research, and can be read here or here.
References
- Bengtsson-Palme J, Gunnarsson L, Larsson DGJ: Can branding and price of pharmaceuticals guide informed choices towards improved pollution control during manufacturing? Journal of Cleaner Production, 171, 137–146 (2018). doi: 10.1016/j.jclepro.2017.09.247
- Hsu A, Alexandre N, Cohen S, Jao P, Khusainova E: 2016 Environmental Performance Index. Yale University, New Haven, CT, USA (2016). http://epi.yale.edu/reports/2016-report
- Transparency International: Corruption Perceptions Index 2014. Transparency International, Berlin, Germany (2014). http://www.transparency.org/cpi2014/in_detail
- Larsson DGJ, Fick J: Transparency throughout the production chain–a way to reduce pollution from the manufacturing of pharmaceuticals? Regulatory Toxicology and Pharmacology, 53, 161–163 (2009). doi:10.1016/j.yrtph.2009.01.008
- Ågerstrand M, Berg C, Björlenius B, Breitholtz M, Brunström B, Fick J, Gunnarsson L, Larsson DGJ, Sumpter JP, Tysklind M, Rudén C: Improving environmental risk assessment of human pharmaceuticals. Environmental Science & Technology, 49, 5336–5345 (2015). doi:10.1021/acs.est.5b00302
- Industry Roadmap for Progress on Combating Antimicrobial Resistance: Industry Roadmap for Progress on Combating Antimicrobial Resistance – September 2016. (2016). http://www.ifpma.org/wp-content/uploads/2016/09/Roadmap-for-Progress-on-AMR-FINAL.pdf
- Larsson DGJ, de Pedro C, Paxeus N: Effluent from drug manufactures contains extremely high levels of pharmaceuticals. Journal of Hazardous Materials, 148, 751–755 (2007). doi:10.1016/j.jhazmat.2007.07.008
- aus der Beek T, Weber FA, Bergmann A, Hickmann S, Ebert I, Hein A, Küster A: Pharmaceuticals in the environment–Global occurrences and perspectives. Environmental Toxicology and Chemistry, 35, 823–835 (2016). doi:10.1002/etc.3339
- Bengtsson-Palme J, Larsson DGJ: Concentrations of antibiotics predicted to select for resistant bacteria: Proposed limits for environmental regulation. Environment International, 86, 140–149 (2016). doi: 10.1016/j.envint.2015.10.015
- Bengtsson-Palme J, Larsson DGJ: Time to limit antibiotic pollution. The Medicine Maker, 0416, 302, 17–18 (2016). [Paper link]
Published paper: Environmental factors leading to resistance
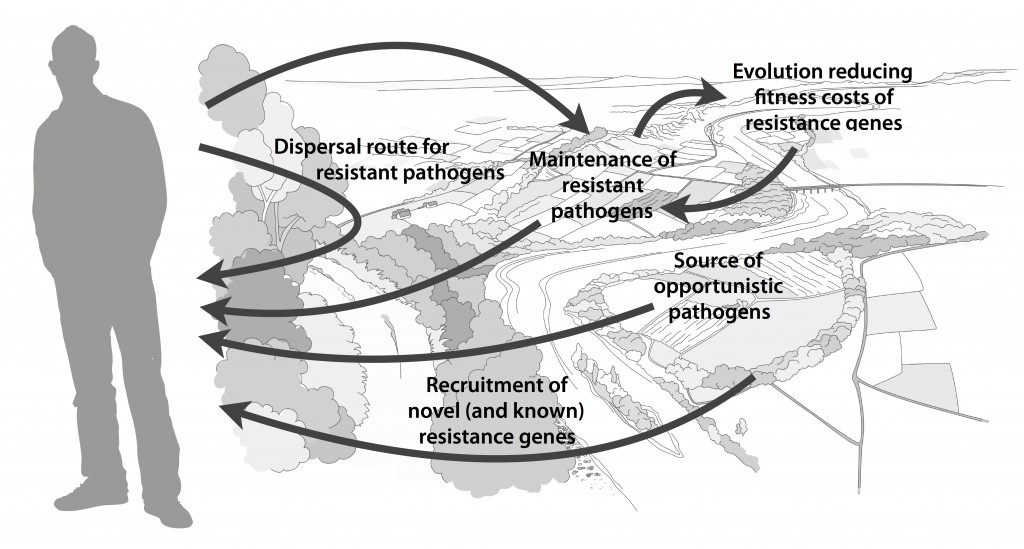
Myself, Joakim Larsson and Erik Kristiansson have written a review on the environmental factors that influence development and spread of antibiotic resistance, which was published today in FEMS Microbiology Reviews. The review (1) builds on thoughts developed in the latter parts of my PhD thesis (2), and seeks to provide a synthesis knowledge gained from different subfields towards the current understanding of evolutionary and ecological processes leading to clinical appearance of resistance genes, as well as the important environmental dispersal barriers preventing spread of resistant pathogens.
We postulate that emergence of novel resistance factors and mobilization of resistance genes are likely to occur continuously in the environment. However, the great majority of such genetic events are unlikely to lead to establishment of novel resistance factors in bacterial populations, unless there is a selection pressure for maintaining them or their fitness costs are negligible. To enable measures to prevent resistance development in the environment, it is therefore critical to investigate under what conditions and to what extent environmental selection for resistance takes place. Selection for resistance is likely less important for the dissemination of resistant bacteria, but will ultimately depend on how well the species or strain in question thrives in the external environment. Metacommunity theory (3,4) suggests that dispersal ability is central to this process, and therefore opportunistic pathogens with their main habitat in the environment may play an important role in the exchange of resistance factors between humans and the environment. Understanding the dispersal barriers hindering this exchange is not only key to evaluate risks, but also to prevent resistant pathogens, as well as novel resistance genes, from reaching humans.
Towards the end of the paper, we suggest certain environments that seem to be more important from a risk management perspective. We also discuss additional problems linked to the development of antibiotic resistance, such as increased evolvability of bacterial genomes (5) and which other types of genes that may be mobilized in the future, should the development continue (1,6). In this review, we also further develop thoughts on the relative risks of re-recruiting and spreading well-known resistance factors already circulating in pathogens, versus recruitment of completely novel resistance genes from environmental bacteria (7). While the latter case is likely to be very rare, and thus almost impossible to quantify the risks for, the consequences of such (potentially one-time) events can be dire.
I personally think that this is one of the best though-through pieces I have ever written, and since it is open access and (in my biased opinion) written in a fairly accessible way, I recommend everyone to read it. It builds on the ecological theories for resistance ecology developed by, among others, Fernando Baquero and José Martinez (8-13). Over the last year, it has been stressed several times at meetings (e.g. at the EDAR conferences in August) that there is a need to develop an ecological framework for antibiotic resistance genes. I think this paper could be one of the foundational pillars on such an endeavor and look forward to see how it will fit into the growing literature on the subject!
References
- Bengtsson-Palme J, Kristiansson E, Larsson DGJ: Environmental factors influencing the development and spread of antibiotic resistance. FEMS Microbiology Reviews, accepted manuscript (2017). doi: 10.1093/femsre/fux053
- Bengtsson-Palme J: Antibiotic resistance in the environment: a contribution from metagenomic studies. Doctoral thesis (medicine), Department of Infectious Diseases, Institute of Biomedicine, Sahlgrenska Academy, University of Gothenburg, 2016. [Link]
- Bengtsson J: Applied (meta)community ecology: diversity and ecosystem services at the intersection of local and regional processes. In: Verhoef HA, Morin PJ (eds.). Community Ecology: Processes, Models, and Applications. Oxford: Oxford University Press, 115–130 (2009).
- Leibold M, Norberg J: Biodiversity in metacommunities: Plankton as complex adaptive systems? Limnology and Oceanography, 1278–1289 (2004).
- Gillings MR, Stokes HW: Are humans increasing bacterial evolvability? Trends in Ecology and Evolution, 27, 346–352 (2012).
- Gillings MR: Evolutionary consequences of antibiotic use for the resistome, mobilome and microbial pangenome. Frontiers in Microbiology, 4, 4 (2013).
- Bengtsson-Palme J, Larsson DGJ: Antibiotic resistance genes in the environment: prioritizing risks. Nature Reviews Microbiology, 13, 369 (2015). doi: 10.1038/nrmicro3399-c1
- Baquero F, Alvarez-Ortega C, Martinez JL: Ecology and evolution of antibiotic resistance. Environmental Microbiology Reports, 1, 469–476 (2009).
- Baquero F, Tedim AP, Coque TM: Antibiotic resistance shaping multi-level population biology of bacteria. Frontiers in Microbiology, 4, 15 (2013).
- Berendonk TU, Manaia CM, Merlin C et al.: Tackling antibiotic resistance: the environmental framework. Nature Reviews Microbiology, 13, 310–317 (2015).
- Hiltunen T, Virta M, Laine A-L: Antibiotic resistance in the wild: an eco-evolutionary perspective. Philosophical Transactions of the Royal Society B: Biological Sciences, 372 (2017) doi: 10.1098/rstb.2016.0039.
- Martinez JL: Bottlenecks in the transferability of antibiotic resistance from natural ecosystems to human bacterial pathogens. Frontiers in Microbiology, 2, 265 (2011).
- Salyers AA, Amábile-Cuevas CF: Why are antibiotic resistance genes so resistant to elimination? Antimicrobial Agents and Chemotherapy, 41, 2321–2325 (1997).
Published paper: 76 new metallo-beta-lactamases
Today, Microbiome put online a paper lead-authored by my colleague Fanny Berglund – one of Erik Kristiansson‘s brilliant PhD students – in which we identify 76 novel metallo-ß-lactamases (1). This feat was made possible because of a new computational method designed by Fanny, which uses a hidden Markov model based on known B1 metallo-ß-lactamases. We analyzed over 10,000 bacterial genomes and plasmids and over 5 terabases of metagenomic data and could thereby predict 76 novel genes. These genes clustered into 59 new families of metallo-β-lactamases (given a 70% identity threshold). We also verified the functionality of 21 of these genes experimentally, and found that 18 were able to hydrolyze imipenem when inserted into Escherichia coli. Two of the novel genes contained atypical zinc-binding motifs in their active sites. Finally, we show that the B1 metallo-β-lactamases can be divided into five major groups based on their phylogenetic origin. It seems that nearly all of the previously characterized mobile B1 β-lactamases we identify in this study were likely to have originated from chromosomal genes present in species within the Proteobacteria, particularly Shewanella spp.
This study more than doubles the number of known B1 metallo-β-lactamases. As with the study by Boulund et al. (2) which we published last month on computational discovery of novel fluoroquinolone resistance genes (which used a very similar approach but on a completely different type of genes), this study also supports the hypothesis that environmental bacterial communities act as sources of uncharacterized antibiotic resistance genes (3-7). Fanny have done a fantastic job on this paper, and I highly recommend reading it in its entirety (it’s open access so you have virtually no excuse not to). It can be found here.
References
- Berglund F, Marathe NP, Österlund T, Bengtsson-Palme J, Kotsakis S, Flach C-F, Larsson DGJ, Kristiansson E: Identification of 76 novel B1 metallo-β-lactamases through large-scale screening of genomic and metagenomic data. Microbiome, 5, 134 (2017). doi: 10.1186/s40168-017-0353-8
- Boulund F, Berglund F, Flach C-F, Bengtsson-Palme J, Marathe NP, Larsson DGJ, Kristiansson E: Computational discovery and functional validation of novel fluoroquinolone resistance genes in public metagenomic data sets. BMC Genomics, 18, 682 (2017). doi: 10.1186/s12864-017-4064-0
- Bengtsson-Palme J, Larsson DGJ: Antibiotic resistance genes in the environment: prioritizing risks. Nature Reviews Microbiology, 13, 369 (2015). doi: 10.1038/nrmicro3399-c1
- Allen HK, Donato J, Wang HH et al.: Call of the wild: antibiotic resistance genes in natural environments. Nature Reviews Microbiology, 8, 251–259 (2010).
- Berendonk TU, Manaia CM, Merlin C et al.: Tackling antibiotic resistance: the environmental framework. Nature Reviews Microbiology, 13, 310–317 (2015).
- Martinez JL: Bottlenecks in the transferability of antibiotic resistance from natural ecosystems to human bacterial pathogens. Frontiers in Microbiology, 2, 265 (2011).
- Finley RL, Collignon P, Larsson DGJ et al.: The scourge of antibiotic resistance: the important role of the environment. Clinical Infectious Diseases, 57, 704–710 (2013).
Published paper: Computational discovery of novel qnr genes
BMC Genomics today published a paper first-authored by my long-time colleague Fredrik Boulund, which describes a computational screen of genomes and metagenomes for novel qnr fluoroquinolone resistance genes (1). The study makes use of Fredrik’s well-designed and updated qnr-prediction pipeline, but in contrast to his previous publication based on the pipeline from 2012 (2), we here study a 20-fold larger dataset of almost 13 terabases of sequence data. Based on this data, the pipeline predicted 611 putative qnr genes, including all previously described plasmid-mediated qnr gene families. 20 of the predicted genes were previously undescribed, and of these nine were selected for experimental validation. Six of those tested genes improved the survivability under ciprofloxacin exposure when expressed in Escherichia coli. The study shows that qnr genes are almost ubiquitous in environmental microbial communities. This study also lends further credibility to the hypothesis that environmental bacterial communities can act as sources of previously uncharacterized antibiotic resistance genes (3-7). The study can be read in its entirety here.
References
- Boulund F, Berglund F, Flach C-F, Bengtsson-Palme J, Marathe NP, Larsson DGJ, Kristiansson E: Computational discovery and functional validation of novel fluoroquinolone resistance genes in public metagenomic data sets. BMC Genomics, 18, 682 (2017). doi: 10.1186/s12864-017-4064-0
- Boulund F, Johnning A, Pereira MB, Larsson DGJ, Kristiansson E: A novel method to discover fluoroquinolone antibiotic resistance (qnr) genes in fragmented nucleotide sequences. BMC Genomics, 13, 695 (2012). doi: 10.1186/1471-2164-13-695
- Bengtsson-Palme J, Larsson DGJ: Antibiotic resistance genes in the environment: prioritizing risks. Nature Reviews Microbiology, 13, 369 (2015). doi: 10.1038/nrmicro3399-c1
- Allen HK, Donato J, Wang HH et al.: Call of the wild: antibiotic resistance genes in natural environments. Nature Reviews Microbiology, 8, 251–259 (2010).
- Berendonk TU, Manaia CM, Merlin C et al.: Tackling antibiotic resistance: the environmental framework. Nature Reviews Microbiology, 13, 310–317 (2015).
- Martinez JL: Bottlenecks in the transferability of antibiotic resistance from natural ecosystems to human bacterial pathogens. Frontiers in Microbiology, 2, 265 (2011).
- Finley RL, Collignon P, Larsson DGJ et al.: The scourge of antibiotic resistance: the important role of the environment. Clinical Infectious Diseases, 57, 704–710 (2013).
Published paper: The Calanus glacialis mitogenome
Mitochondrial DNA Part B today published a mitochondrial genome announcement paper (1) in which I was involved in doing the assemblies and annotating them. The paper describes the mitogenome of Calanus glacialis, a marine planktonic copepod, which is a keystone species in the Arctic Ocean. The mitogenome is 20,674 bp long, and includes 13 protein-coding genes, 2 rRNA genes and 22 tRNA genes. While this is of course note a huge paper, we believe that this new resource will be of interest in understanding the structure and dynamics of C. glacialis populations. The main work in this paper has been carried out by Marvin Choquet at Nord University in Bodø, Norway. So hats off to him for great work, thanks Marvin! The paper can be read here.
Reference
- Choquet M, Alves Monteiro HJ, Bengtsson-Palme J, Hoarau G: The complete mitochondrial genome of the copepod Calanus glacialis. Mitochondrial DNA Part B, 2, 2, 506–507 (2017). doi: 10.1080/23802359.2017.1361357 [Paper link]
Published paper: Investigating resistomes using metagenomics
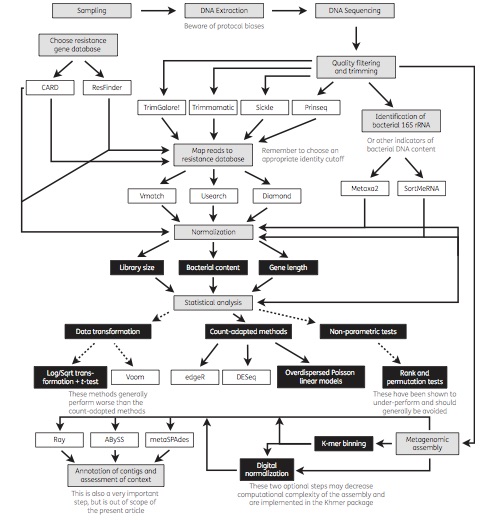
Today, a review paper which I wrote together with Joakim Larsson and Erik Kristiansson was published in Journal of Antimicrobial Chemotherapy (1). We have for a long time used metagenomic DNA sequencing to study antibiotic resistance in different environments (2-6), including in the human microbiota (7). Generally, our ultimate purpose has been to assess the risks to human health associated with resistance genes in the environment. However, a multitude of methods exist for metagenomic data analysis, and over the years we have learned that not all methods are suitable for the investigation of resistance genes for this purpose. In our review paper, we describe and discuss current methods for sequence handling, mapping to databases of resistance genes, statistical analysis and metagenomic assembly. We also provide an overview of important considerations related to the analysis of resistance genes, and end by recommending some of the currently used tools, databases and methods that are best equipped to inform research and clinical practice related to antibiotic resistance (see the figure from the paper below). We hope that the paper will be useful to researchers and clinicians interested in using metagenomic sequencing to better understand the resistance genes present in environmental and human-associated microbial communities.
References
- Bengtsson-Palme J, Larsson DGJ, Kristiansson E: Using metagenomics to investigate human and environmental resistomes. Journal of Antimicrobial Chemotherapy, advance access (2017). doi: 10.1093/jac/dkx199 [Paper link]
- Bengtsson-Palme J, Boulund F, Fick J, Kristiansson E, Larsson DGJ: Shotgun metagenomics reveals a wide array of antibiotic resistance genes and mobile elements in a polluted lake in India. Frontiers in Microbiology, 5, 648 (2014). doi: 10.3389/fmicb.2014.00648 [Paper link]
- Lundström S, Östman M, Bengtsson-Palme J, Rutgersson C, Thoudal M, Sircar T, Blanck H, Eriksson KM, Tysklind M, Flach C-F, Larsson DGJ: Minimal selective concentrations of tetracycline in complex aquatic bacterial biofilms. Science of the Total Environment, 553, 587–595 (2016). doi: 10.1016/j.scitotenv.2016.02.103 [Paper link]
- Bengtsson-Palme J, Hammarén R, Pal C, Östman M, Björlenius B, Flach C-F, Kristiansson E, Fick J, Tysklind M, Larsson DGJ: Elucidating selection processes for antibiotic resistance in sewage treatment plants using metagenomics. Science of the Total Environment, 572, 697–712 (2016). doi: 10.1016/j.scitotenv.2016.06.228 [Paper link]
- Pal C, Bengtsson-Palme J, Kristiansson E, Larsson DGJ: The structure and diversity of human, animal and environmental resistomes. Microbiome, 4, 54 (2016). doi: 10.1186/s40168-016-0199-5 [Paper link]
- Flach C-F, Pal C, Svensson CJ, Kristiansson E, Östman M, Bengtsson-Palme J, Tysklind M, Larsson DGJ: Does antifouling paint select for antibiotic resistance? Science of the Total Environment, 590–591, 461–468 (2017). doi: 10.1016/j.scitotenv.2017.01.213 [Paper link]
- Bengtsson-Palme J, Angelin M, Huss M, Kjellqvist S, Kristiansson E, Palmgren H, Larsson DGJ, Johansson A: The human gut microbiome as a transporter of antibiotic resistance genes between continents. Antimicrobial Agents and Chemotherapy, 59, 10, 6551–6560 (2015). doi: 10.1128/AAC.00933-15 [Paper link]
Talk on emission limits in Stockholm
In two weeks time, on the 15th of June, I will participate in a seminar organised by Landstingens nätverk för läkemedel och miljö (the Swedish county council network for pharmaceuticals and environment; the seminar will be held in Swedish) in Stockholm. I will give a talk on our proposed emission limits for antibiotics published last year (the paper is available here), but there will also be talks on wastewater treatment, sustainable pharmaceutical usage and environmental standards for pharmaceuticals. The full program can be found here, and you may register here until June 9. The seminar is free of charge.
And if you are interested in this, I can also recommend the webinar given by Healthcare Without Harm next week (on June 8), which will deal with sustainable procurement as a means to deal with pharmaceutical pollution in the environment. I will at least tune in to hear how the discussion goes here.
Report on JRC AMR workshop
In March, I attended a workshop on the role of NGS technologies in the coordinated action plan against antimicrobial resistance, organised by JRC in Italy. I was, together with 14 other experts, invited to discuss where and how sequencing can be used to investigate and manage antibiotic resistance. The report from the workshop has just recently been published, and is available here. There will be follow-up activities on this workshop, which I also hope that I will be able to participate in, since this is an important and very interesting pet topic of mine.
Reference