My ISME talk on EMBARK
Ákos Kovács had the brilliant idea of putting up a temporary resource for things you bring up in a talk that you can point people to. I did not do this before my talk at ISME today, but I thought the idea was so good, so here’s a summary and collection of my ISME short-talk on the EMBARK outcomes today:
- More information on EMBARK and its successor SEARCHER can be found on the project website, here: http://antimicrobialresistance.eu Importantly, this is a team effort over four years and I only touched on a few selected things
- Within the project we have looked at typical background levels of antibiotic resistance in the environment. We have already published some of these results (for qPCR abundances) in Abramova et al. 2023
- The average resistance gene in the average environment is present in ~1 in 1000 bacteria, but the variation between different genes is huge
- Depending on monitoring goal, different target genes are relevant to use. See this table adapted from Abramova et al. 2023:
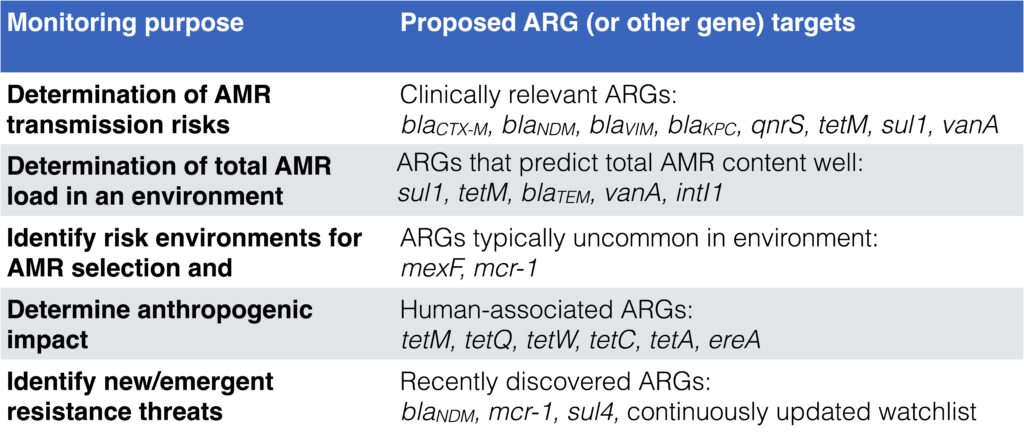
- We have also tried to make different monitoring methods for environmental AMR comparable. Those mentioned in the talk were selective culturing for resistant bacteria, qPCR and shotgun metagenomics
- This data is not yet published, but overall we see relatively good correlation between qPCR and metagenomics. This is not true for all genes, though, and unfortunately neither qPCR nor metagenomics is always better than the other
- Culturing data is not very good at predicting specific antibiotic resistance gene abundances as the class level
- Finally, we have developed methods for discovering new types of ARGs, as seen in the ResFinderFG database: Gschwind et al. 2023
- We have also used these new methods to look at differences between established ARGs and latent ARGs in a variety of environments: Inda-Díaz et al. 2023
- Our ultimate goal in EMBARK would be to develop a modular framework for environmental monitoring of antibiotic resistance. You can read more about our thinking and goals in the review paper we published last year: Bengtsson-Palme et al. 2023
Heading to Seoul for ISME15
 Finally, I am at the airport waiting for the plane to Beijing, and tomorrow, the plane to Seoul. I will spend next week at the ISME15 conference, which will be awesome. I hope to meet as many of you there as possible!
Finally, I am at the airport waiting for the plane to Beijing, and tomorrow, the plane to Seoul. I will spend next week at the ISME15 conference, which will be awesome. I hope to meet as many of you there as possible!
(And on a side note, if I don’t answer mails it might be due to that the conference wifi might be overcrowded. They warned us about this potential shortcoming of the conference center…)
My ISME Program for Thursday and Friday
So I finally have finalized my own program for what I’m going to see at ISME today and tomorrow. As the last time, the things I think I will actually attend have bold times. Generally, thursday is easy – I will be at the Bioinformatics session, where I will also present myself (at 13.30 in Hall A1). Friday is a little bit trickier, but considerably less complicated to choose from than tuesday was. See you around, and don’t forget to visit my poster (board 002A)!
Thursday
Morning session
08:30 – 09:20 From omics to the environment and back: unraveling how chemosynthetic symbionts gain energy and carbon
Nicole Dubilier, Symbiosis Group, Max Planck Institute of Marine Microbiology, Bremen, Germany (Hall A1)
1000 – 1030 Bayesian hierarchical models for defining enterotypes and ecotypes
Christopher Quince, University of Glasgow, UK (Hall A1)
1000 – 1030 Composition, structure and function of hot spring cyanobacterial mat communities: use of high-throughput technologies and theory to demarcate guilds, species and the functions they catalyze
Dave Ward, Montana State University, Bozeman, USA (Hall A2)
1000 – 1030 Stability and Resilience in the Human Microbiome
David Relman, Stanford University, USA (Auditorium 11)
1030 – 1100 From the HMP to the EMP: deriving insight from large-scale sequencing projects
Rob Knight, University of Colorado, USA (Hall A1)
1100 – 1130 Phylogenetic conservatism of functional traits in microorganisms
Adam Martiny, University of California, Irvine, USA (Hall A1)
1130 – 1200 Title to be confirmed
Jeroen Raes, VIB, Belgium (Hall A1)
1130 – 1200 Feast then famine: Fe bioavailability and utilization through geological time
Christopher Dupont, J. Craig Venter Institute, USA (Auditorium 15)
Lunch session
12:30 – 13:15 Tackling the pitfalls and unveiling the promise of soil metagenomics
Janet Jansson, Earth Sciences Division, Lawrence Berkeley, National Laboratory, Berkeley, CA, USA (Hall A1)
Afternoon session
1330 – 1345 Comprehensive Analysis of Antibiotic Resistance Genes in River Sediment, Well Water and Soil Microbial Communities Using Metagenomic DNA Sequencing
Johan Bengtsson [Sweden] (Hall A1)
1330 – 1345 Genome-wide diversification patterns of fresh and saltwater bacteria of the SAR11 clade inferred from single cell and metagenomics data
Siv Andersson [Sweden] (Auditorium 15)
1330 – 1345 Spatial and functional organization of deep-water microbial communities in the ocean: role of particle attachment
Gerhard J. Herndl [Austria] (Session Room B4)
1345 – 1400 Investigating diversified gene functions in microbial communities of the human gut
Lina Faller [USA] (Hall A1)
1400 – 1415 Large-scale characterization of the diversity and community composition of human intestinal microbiota
Leo Lahti [Netherlands] (Hall A1)
1400 – 1415 Significant and persistent impact of timber harvesting on soil microbial communities in northern coniferous forests
Martin Hartmann [Switzerland] (Hall A3)
1400 – 1415 From communities to single cells: finding functional coherence in the lake microbiome
Stefan Bertilsson [Sweden] (Session Room B4)
1415 – 1430 Deciphering the microbial community and the lignocellulolytic digestome of lower termite Coptotermes gestroi
João Paulo Franco Cairo [Brazil] (Hall A1)
1415 – 1430 Polyextremotolerant human opportunistic black yeasts inhabit dishwashers around the world
Nina Gunde-Cimerman [Slovenia] (Hall A2)
1415 – 1430 Dating the cyanobacterial origin of the chloroplast
Luisa Falcon [Mexico] (Auditorium 15)
1430 – 1445 Linking genotypes to phenotypes: characterizing viral proteins of unknown function
Jeremy Frank [USA] (Hall A1)
1430 – 1445 Living together apart: soil metagenomics and microbially relevant scales
George A. Kowalchuk [Netherlands] (Session Room B4)
1445 – 1500 The metagenomics of the dead: taxonomic and functional annotation methods for analysis of ancient datasets
Katarzyna Zaremba-Niedzwiedzka [Sweden] (Hall A1)
1445 – 1500 Ecological drivers of bacterial social evolution
Ines Mandic Mulec [Slovenia] (Auditorium 15)
1500 – 1515 Dirty little secrets for soil metagenomic assembly
Adina Howe [USA] (Hall A1)
1500 – 1515 Antibiotic resistance associated with waste water treatment plant effluent
Gregory Amos [United Kingdom] (Session Room B3)
1515 – 1530 Application of Unifrac and related bioinformatic tools to assess intra-species diversity within oral microbiomes in an anthropological context
Hans-Peter Horz [Germany] (Hall A1)
1515 – 1530 Comparative genomics of the ubiquitous, hydrocarbon degrading genus Marinobacter
Esther Singer [USA] (Auditorium 15)
Friday
Morning session
1000 – 1030 The fate of pesticides in soil and aquifers from a small-scale point of view: Does microbial and spatial heterogeneity have an impact?
Jens Aamand, The Geological Survey of Denmark and Greenland (GEUS), Denmark (Hall A2)
1000 – 1030 Commonness and rarity: Core and satellite species groups in microbial metacommunities
Christopher van der Gast, NERC Centre for Ecology & Hydrology, Wallingford, UK (Auditorium 11)
1100 – 1130 Spatial heterogeneity at the microbial scale: effects on microbial community structure and functioning
Naoise Nunan, BioEMCo, Centre INRA Versailles-Grignon, France (Hall A2)
1100 – 1130 Signalling controlled structure function of activated sludge microbial communities
Staffan Kjelleberg, University of New South Wales, Australia (Auditorum 15)
1130 – 1200 The impact of soil spatial heterogeneity on concepts of microbial interactions and diversity
James I. Prosser, University of Aberdeen, UK (Hall A2)
1130 – 1200 Micro-scale spatial expansion of microbial cells and mobile genetic elements
Barth Smets, Technical University of Denmark, Denmark (Auditorium 11)
Afternoon session
1400 – 1415 Extracellular vesicles from the marine cyanobacterium Prochlorococcus may mediate diverse community interactions
Steven Biller [USA] (Hall A1)
1415 – 1430 Simultaneous amplicon sequencing to explore co-occurrence patterns of bacterial, archaeal and eukaryotic components of rumen microbial communities
Sandra Kittelmann [New Zealand] (Session Room B4)
1430 – 1445 The enterotypes of great ape gut microbiomes
Andrew Moeller [USA] (Session Room B4)
1445 – 1500 Theoretical models for bacterial communities in drinking water as they travel and evolve through drinking water distribution systems
Joanna Schroeder [United Kingdom] (Auditorium 11)
1500 – 1515 Wastewater treatment plants from across the globe have a reproducible core microbial community
Aaron Marc Saunders Denmark] (Session Room B4)
1530 – 1545 Every cloud has a silver lining: invasion by Limnohabitans planktonicus promotes the maintenance of diversity in bacterial communities
Gianluca Corno [Italy] (Hall A1)
1530 – 1545 Metabolic flexibility as a major predictor of spatial distribution in microbial communities
Kevin Purdy [United Kingdom] (Auditorium 15)
1545 – 1600 Microbial dynamics in a thawing world: linking microbial communities to increased methane flux in degrading permafrost
Gene Tyson [Australia] (Auditorium 15)
My ISME Program Monday and Tuesday
I have tried to put together a list of talks I find interesting at ISME14 next week (oh, well going through the program book I find virtually all talks interesting, but some of them more so than others). I didn’t get there quite yet, but I have a list for monday and tuesday. I have realized that monday morning will likely run smoothly (a tough call between von Mering and Gilbert at 11.30, but otherwise the choices were not too hard). Monday afternoon I will probably spend in Session Room B3, where the Microbial Community Diversity: 16S and Beyond session will take place (many sessions have really silly, non-informative, names at this conference – what would not fit into the concept “16S and beyond“?). I will really miss the talk by Otto X. Cordero on bacteria act as socially cohesive units and antibiotic resistance, but I simply don’t think there will be time for a trip to Hall A2 and back just for that talk.
Another tough call is the round table discussions on Monday evening. Here, however, I feel that I have a duty to attend to the Unraveling the bacterial mobilome: Potentials and limitations of the present methodology session in Auditorium 11, which also might be the most relevant one for my current research projects.
Below you find my list of interesting talks at ISME14, sorted by time. Times in bold are the ones I will aim to attend in case I have to make a choice. Also, of course I will attend my own poster presentation on Monday afternoon (poster board number 267A). Hope to see you there!
Monday
Morning session
1000 – 1030 Cross-biome comparisons of soil microbial communities and their functional potentials
Noah Fierer, University of Colorado at Boulder, USA (Hall A2)
1030 – 1100 Experimental biogeography of bacteria in miniature ecosystems
Thomas Bell, Imperial College, London, UK (Hall A2)
1130 – 1200 Tracking OTUs around the environment – a challenge for clustering algorithms and interpretation
Christian von Mering, University of Zurich, Switzerland (Hall A1)
1130 – 1200 The Earth Microbiome Project: A new paradigm in geospatial and temporalstudies of microbial ecology
Jack A. Gilbert, Argonne National Laboratory and University of Chicago, USA (Hall A2)
Afternoon session
1330 – 1345 What makes a bacterium fresh? Genome wide functional comparison of marine and freshwater SAR11
Alexander Eiler [Sweden] (Auditorium 15)
1330 – 1345 Rarity and the problem of measuring diversity
Bart Haegeman [France] (Session Room B3)
1345 – 1400 Complexity does not necessarily create diversity
Tom Curtis [United Kingdom] (Session Room B3)
1345 – 1400 Depletion of the rare bacterial biosphere in expanding oceanic oxygen minimum zones
J. Michael Beman [USA] (Session Room B4)
1400 – 1415 Selection history affects the predictability of microbial ecosystem development
Andrew Free [United Kingdom] (Session Room B3)
1415 – 1430 Can we use microbial life strategies to understand the response of microbial communities to moisture stress?
Sarah Evans [USA] (Session Room B3)
1430 – 1445 Towards a unified taxonomy for ribosomal RNA databases
Pelin Yilmaz [Germany] (Session Room B3)
1445 – 1500 Systematic design of 18S rDNA primers for assessing eukaryotic diversity
Luisa Hugerth [Sweden] (Session Room B3)
1445 – 1500 Either Of yeast, grapes and wasps: Saccharomyces cerevisiae ecology revised
Irene Stefanini [Italy] (Hall A1)
1500 – 1515 Soil bacterial biogeography: a taxonomic and functional perspective
Rob Griffiths [United Kingdom] (Hall A1)
1500 – 1515 Ecological populations of bacteria act as socially cohesive units of antibiotic production and resistance in the wild
Otto X. Cordero [USA] (Hall A2)
1515 – 1530 Micro-scale drivers of bacterial diversity and biogeography
George Kowalchuk [Netherlands] (Hall A1)
1515 – 1530 Time and space resolved deep metagenomics to investigate selection pressures on low abundant species in complex environments
Mads Albertsen [Denmark] (Session Room B3)
1515 – 1530 Bacterial community structure and composition in high arsenic contaminated ground water from West Bengal and microbial role in subsurface arsenic release
Pinaki Sar [India] (Auditorium 10)
Evening session (round-table discussions)
17:30 – 19:30 RT11: Microbial Network Ecology: Deciphering Complex Network Interactions in Microbial Communities (Hall A3)
17:30 – 19:30 RT16: Unraveling the bacterial mobilome: Potentials and limitations of the present methodology (Auditorium 11)
17:30 – 19:30 RT17: Microbial invasions: What defines whether a microorganism is an invasive species? (Auditorium 12)
Tuesday
Morning session
08:30 – 09:20 You are what you eat but not always what omics predicts: On the importance of single cell ecophysiology of microbes
Michael Wagner, Department of Microbial Ecology, University of Vienna, Austria (Hall A1)
1000 – 1030 Can dormancy theory help us retrieve rare and uncultured microbes?
Jay Lennon, Indiana University, USA (Hall A3)
1000 – 1030 A critical review of mean metabolic rates of subsurface microbial communities
Bo Barker Jørgensen, University of Aarhus, Denmark (Auditorum 11)
1000 – 1030 Horizontal genetic transfer and the origin of species in bacteria
Frederick M. Cohan, Wesleyan University, USA (Auditorum 12)
1030 – 1100 Phylogenomic networks reveal mechanisms for lateral gene transfer during microbial evolution
Tal Dagan, Heinrich Heine University Düsseldorf, Germany (Auditorum 12)
1100 – 1130 Coevolution between plasmids and their hosts: consequences for the persistence of drug resistance
Eva Top, University of Idaho, USA (Auditorum 12)
1130 – 1200 The communal gene pool in natural environments
Søren J. Sørensen, University of Copenhagen, Denmark (Auditorum 12)
1130 – 1200 Let genomes do the talking: Using genomics to unravel genes under selection in archaeal populations
Rachel Whitaker, University of Illinois, USA (Auditorum 15)
Afternoon session
1330 – 1345 Spatial patterns of microbial communities at a soil microscale
Florentin Constancias [France] (Hall A2)
1330 – 1345 Microbial biofilm biodiversity distribution in a stream network
Katharina Besemer [Austria] (Session Room B3)
1330 – 1345 Effects of loss of rare microbes on soil ecosystem services
Gera Hol [Netherlands] (Auditorium 10)
1345 – 1400 Targeted recovery of novel phylogenetic diversity from next-generation sequence data
Josh D. Neufeld [Canada] (Auditorium 10)
1345 – 1400 Functionally relevant microdiversity of Nitrospira-like bacteria in activated sludge
Christiane Dorninger [Austria] (Hall A3)
1345 – 1400 Hot spot of horizontal gene transfer: high abundance and diversity of mobile genetic elements in bacterial communities of on-farm pesticide biopurification systems
Kornelia Smalla [Germany] (Auditorium 12)
1400 – 1415 Transcriptional dynamics of catabolic genes in soil – fine-scale analysis for a deeper understanding of soil functioning
Mette H Nicolaisen [Denmark] (Hall A1)
1400 – 1415 Seasonal synchronicity and specificity of microbial community and population dynamics across four alpine lakes
Ryan Mueller [Spain] (Session Room B3)
1400 – 1415 Mapping genotypic diversity onto niche adaptation
Yutaka Yawata [USA] (Auditorium 12)
1415 – 1430 Soil bacterial community shift after chitin enrichment: an integrative metagenomic approach
Samuel Jacquiod [France] (Hall A3)
1415 – 1430 MetaGenomic Species: adding structure to metagenomics data
H. Bjørn Nielsen [Denmark] (Auditorium 12)
1430 – 1445 Impact of diet on human gut microbial communities
Cindy Nakatsu [USA] (Hall A3)
1430 – 1445 Insights into the bovine rumen plasmidome
Itzhak Mizrahi [Israel] (Auditorium 12)
1445 – 1500 Mechanisms of biocide-induced antibiotic resistance: from single cell to community
Seungdae Oh [USA] (Hall A3)
1445 – 1500 Antibiotic resistance in soil bacterial communities: comparison of metals and antibiotic residues as selecting agents and spatial heterogeneity of coselected resistance patterns
Kristian Koefoed Brandt [Denmark] (Hall A2)
1445 – 1500 Viruses, a controlling force on Prokaryotic diversity?
Ruth-Anne Sandaa [Norway] (Session Room B4)
1445 – 1500 Regulation of transfer of the ICEclc element of Pseudomonas
Jan Roelof van der Meer [Switzerland] (Auditorium 12)
1500 – 1515 Pathogen removal in slow sand filters as revealed by stable isotope probing coupled with next generation sequencing
Sarah Haig [United Kingdom] (Hall A3)
1500 – 1515 Development of zinc tolerance by the ammonia oxidising community is restricted to ammonia oxidising Bacteria, rather than Archaea
Stefan Ruyters [Belgium] (Hall A2)
1500 – 1515 Survival strategies of plasmids in chemostats and biofilms: effect of hostrange and competition
Jan-Ulrich Kreft [United Kingdom] (Auditorium 12)
1515 – 1530 Evaluating process-related and seasonal changes in bacterial community in drinking water treatment and distribution systems
Ameet Pinto [USA] (Hall A3)
1515 – 1530 Distribution, diversity, and evolution of cyclic peptide secondary metabolites in natural populations of marine picocyanobacteria
Andres Cubillos-Ruiz [USA] (Hall A1)
1515 – 1530 Permissiveness of soil microbial communities toward receipt of mobile genetic elements
Sanin Musovic [Denmark] (Auditorium 12)
Evening session
1830 Víctor de Lorenzo, Molecular Environmental Microbiology Laboratory, the Spanish National Research Council, Spain
Conflict management and division of labor in bacterial populations degrading recacitrant aromatics
1915 Stephen J. Giovannoni, Oregon State University, USA
Outliers: Extreme Selection for Minimalism in Ocean Microbial Plankton
ISME14 begins today
I am on my way to Copenhagen for the ISME14 conference that begins today. I’m myself quite excited about this event, and will present three posters (two as first author), and give a short talk on antibiotic resistance gene identification and metagenomics. My talk will be in the Bioinformatics in Microbial Ecology session on Thursday afternoon (at 13.30).
 If you’d like to talk about Metaxa and Megraft, I will present an SSU-oriented poster in the Monday afternoon poster section (board number 267A). My antibiotic resistance gene poster will be presented on Thursday afternoon (board number 002A), and I really encourage everyone interested in metagenomics (especially metagenomic assembly) to come talk to me then! Finally, I am also partially responsible for a poster on periphyton metagenomics with Martin Eriksson as its main author. This poster is also presented on Monday, in the Microbial Dispersion and Biogeography session (board number 021A).
If you’d like to talk about Metaxa and Megraft, I will present an SSU-oriented poster in the Monday afternoon poster section (board number 267A). My antibiotic resistance gene poster will be presented on Thursday afternoon (board number 002A), and I really encourage everyone interested in metagenomics (especially metagenomic assembly) to come talk to me then! Finally, I am also partially responsible for a poster on periphyton metagenomics with Martin Eriksson as its main author. This poster is also presented on Monday, in the Microbial Dispersion and Biogeography session (board number 021A).
I hope to be able to make another post later tonight on what are the “essential” sessions for me on this conference. Hope to see you there soon!