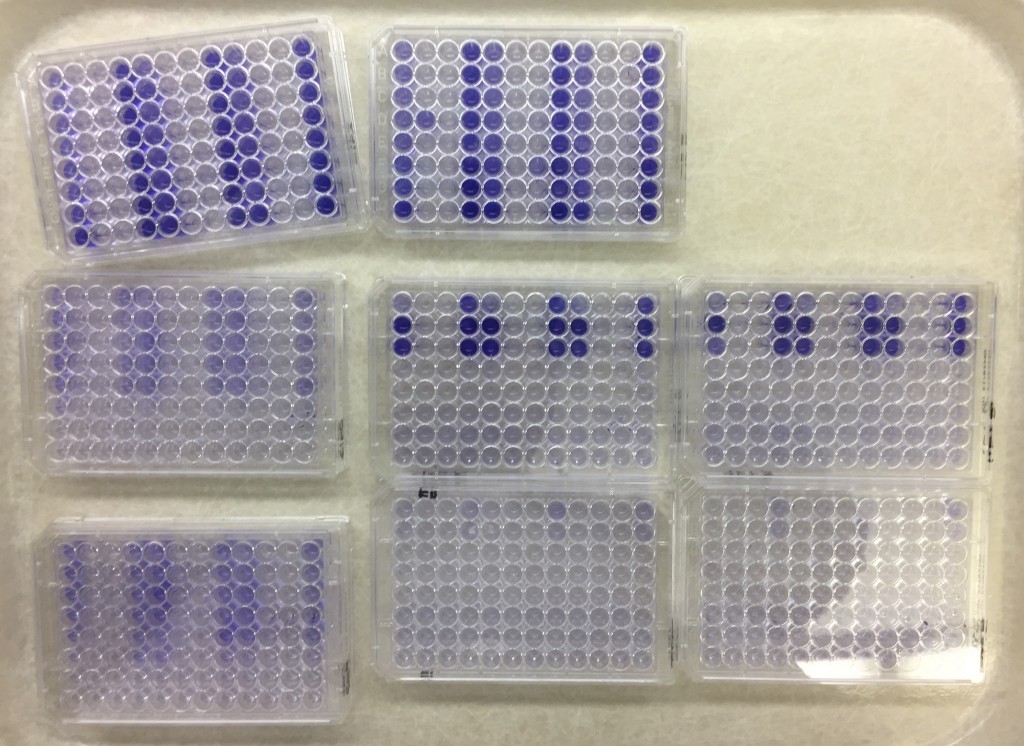
Published paper: Temperature affects community interactions
I am very happy to announce that Emil Burman‘s (doctoral student in the lab) first first-author paper was published today in Frontiers in Microbiology. In this paper (1), we explored how temperature affected the interactions in the model microbial community THOR (2). Somewhat surprisingly, we found that even a small difference in temperature changed the community intrinsic properties (3) of this model community a lot. We furthermore find that changes in growth rates of the members of the community partially explains the changed interaction patterns, but only to some extent. Finally, we also found that biofilm production overall was much higher at lower temperatures (9-15°C) than at room temperature, and that at around 25°C and above the community formed virtually no biofilm.
The temperature range we tested is not unlikely to be encountered when incubating the community in a thermally unregulated environment. Thus, our results show that a high degree of temperature control is crucial between experiments, particularly when reproducing results across different laboratories, equipment, and personnel. This highlights the need for standards and transparency in research on microbial model communities (4).
Another important, related, aspect is that disruptive factors that discriminate against single members of the community are not unique to THOR. Instead, this is likely to be the case for other microbial model (as well as natural communities). Since only a few of these model communities have been elucidated for community behaviors outside of specific culturing conditions they were first contrived under, this may severely limit our view of interactions between microbes to specific laboratory settings. This casts some doubt on the validity of extrapolation from results obtained from microbial model communities. It seems to be important moving forward to establish that community-intrinsic behaviors in model communities are stable in the face of variable environmental conditions, such as temperature, pH, nutrient availability, and initial inoculum size.
A short backstory to this paper: this begun when Emil could not consistently replicate the results I had obtained during my postdoc (working on THOR) in Prof. Jo Handelsman’s lab at the University of Wisconsin-Madison. After a long time of troubleshooting, we realized that our lab did not hold a stable room temperature. We bought a cold incubator, and – boom – after that the expected community behavior came back. This made us realize the importance of temperature for the community-intrinsic properties of THOR, which then led to this more systematic investigation.
Great work Emil! It is nice to finally see this in its published form. Read the entire paper (open access) here!
References
- Burman E, Bengtsson-Palme J: Microbial community interactions are sensitive to small differences in temperature. Frontiers in Microbiology, 12, 672910 (2021). doi: 10.3389/fmicb.2021.672910
- Lozano GL, Bravo JI, Garavito Diago MF, Park HB, Hurley A, Peterson SB, Stabb EV, Crawford JM, Broderick NA, Handelsman J: Introducing THOR, a Model Microbiome for Genetic Dissection of Community Behavior. mBio, 10, 2, e02846-18 (2019). doi: 10.1128/mBio.02846-18
- Madsen JS, Sørensen SJ, Burmølle M: Bacterial social interactions and the emergence of community-intrinsic properties. Current Opinion in Microbiology 42, 104–109 (2018). doi: 10.1016/j.mib.2017.11.018
- Bengtsson-Palme J: Microbial model communities: To understand complexity, harness the power of simplicity. Computational and Structural Biotechnology Journal, 18, 3987-4001 (2020). doi: 10.1016/j.csbj.2020.11.043
Published paper: Microbial model communities
This week, in a stroke of luck coinciding with my conference presentation on the same topic, my review paper on microbial model communities came out in Computational and Structural Biotechnology Journal. The paper (1) provides an overview of the existing microbial model communities that have been developed for different purposes and makes some recommendations on when to use what kind of community. I also make a deep-dive into community intrinsic-properties and how to capture and understand how microbes growing together interact in a way that is not predictable from how they grow in isolation.
The main take-home messages of the paper are that 1) there already exists a quite diverse range of microbial model communities – we probably don’t need a wealth of additional model systems, 2) there need to be better standardization and description of the exact protocols used – this is more important in multi-species communities than when species are grown in isolation, and 3) the researchers working with microbial model communities need to settle on a ‘gold standard’ set of model communities, as well as common definitions, terms and frameworks, or the complexity of the universe of model systems itself may throw a wrench into the research made using these model systems.
The paper was inspired by the work I did in Jo Handelsman‘s lab on the THOR model community (2), which I then have brought with me to the University of Gothenburg. In the lab, we are also setting up other model systems for microbial interactions, and in this process I thought it would be useful to make an overview of what is already out there. And that overview then became this review paper.
The paper is fully open-access, so there is really not much need to go into the details here. Go and read the entire thing instead (or just get baffled by Table 1, listing the communities that are already out there!)
References
- Bengtsson-Palme J: Microbial model communities: To understand complexity, harness the power of simplicity. Computational and Structural Biotechnology Journal, in press (2020). doi: 10.1016/j.csbj.2020.11.043
- Lozano GL, Bravo JI, Garavito Diago MF, Park HB, Hurley A, Peterson SB, Stabb EV, Crawford JM, Broderick NA, Handelsman J: Introducing THOR, a Model Microbiome for Genetic Dissection of Community Behavior. mBio, 10, 2, e02846-18 (2019). doi: 10.1128/mBio.02846-18
Funding from the research council!
I am very happy to share the news that our starting grant application to the Swedish Research Council has been granted 3.3 million SEK of funding for four years! This is fantastic news, as it allows us to further explore the interactions between bacteria in the human microbiome that are important for community stability and resilience to being colonized by pathogens. In the granted project, we will investigate environmental and genetic factors that are important for bacterial invasiveness and community stability in the human gastrointestinal tract.
Within the scope of the project, we will establish model bacterial communities and experimental systems for the human stomach and intestine. We will then investigate how disturbances, such as antibiotic exposure, change the interactions in these microbial communities and their long-term stability. Finally, we aim to identify genes that contribute to successful bacterial colonization or resilience to invasion of established communities in the human microbiome.
Aside from myself, Prof. Sara Lindén and Dr. Kaisa Thorell from the University of Gothenburg as well as Prof. Ed Moore at the university’s Culture Collection will be involved in this project in different ways. We will also collaborate with my former postdoc supervisor Prof. Jo Handelsman as well as Dr. Ophelia Venturelli at the University of Wisconsin-Madison. Finally, we will also collaborate with Dr. Åsa Sjöling at the Karolinska Institute. I look forward to work with you all over the coming four years! A big thanks to the Swedish Research Council for believing in this research and investing in making it happen!
Grant successes!
I am a bit late on the ball here, but January was a great month for the lab in terms of funding. First, we got awarded an Sahlgrenska Academy International Starting Grant – a faculty grant for young researchers comprising of 1 million SEK, intended to support the overall research plan for the lab.
The second grant was awarded by the Centre for Antibiotic Resistance Research (CARe) at the University of Gothenburg and is a project grant focusing on opportunistic pathogens and their role in the emergence and transmission of antibiotic resistance. For this project, we got almost 600,000 SEK over two years to investigate how genes enhancing invasion ability and virulence interact with selection for antibiotic resistance in opportunistic pathogens. The project is somewhat related to the work I did in Prof. Jo Handelsman‘s lab, but extends it to more mechanistic details about how these phenomena are interconnected.
Goodbye Adriana and Welcome Emil
The shift to October marked the last days that our visiting doctoral student Adriana Osińska spent in the lab. Adriana was working on the sequencing data generated from the invasion experiments I performed in Jo Handelsman’s lab. She managed to dig out a great number of genes that seems to have an influence on bacterial community invasion success. Those genes are now candidate genes that will be tested in follow-up studies, which brings us to….
That I forgot to introduce our newest lab member – Emil Burman! Emil is a master student performing his thesis project in the lab and will stay with us until May 2020. Emil will work on experimentally characterizing the candidate genes that Adriana has identified. We are excited to have Emil in the lab and think that he has been off to a great start already. Welcome Emil!
Adriana will no go back to Poland to complete here PhD thesis early next year. We have loved to have her in the lab and she has contributed with data and analyses of tremendous value. We wish her all the best of luck with defending her thesis!
And the experiments have started!
It’s been a long time before I have written something here, mostly because making ourselves at home in Madison have take some time; then we go the flue; and then there have been a lot at work after that. But now i will try to have another go at writing somethings on the Wisconsin Blog. First, a look at my (already messy) desk here at the Wisconsin Institute for Discovery.
And then, a look at my lab space, which I have a view of straight from my desk, through a glass window.
This week, I have started experiments with exposing our little model community to antibiotics and it looks like I’m getting potentially exciting results. I have to sit down with the data today to see if there’s statistical differences, but from the looks of the biofilms, there is potential here.
Next week I will try to start experiment with sand columns and see if I can replicate some of this in this setting as well. It is interesting being back in the lab, and I feel that this an experience that will be very valuable for me going forward. I look forward to the days later this spring when I will start generating sequence data from my own experiments!
Leaving Madison



My first day in Madison


New employment – same work
Today, I started my new position at the University of Gothenburg as a non-tenured assistant professor (forskarassistent)*. In essence, this means that I have a position funded by my own grant until the end of 2020, although I will be on a leave-of-absence while doing my PostDoc with Jo Handelsman in Wisconsin. Speaking of which, I will be leaving to the US on Thursday next week for a month of setting things up at her lab (and also going to the EDAR4 conference in Lansing). I will return to Sweden in mid-September and leave for the US for real early next year.
In terms of actual work, this change of position will not mean very much at the moment. I will continue to do the same things for some time, and I will remain closely associated with Joakim Larsson’s lab at the Dept. of Infectious Diseases. And luckily, I will retain my lovely roommates for at least the time being. In the long run, however, this means that I will shift my research focus slightly, away from antibiotic resistance risk management towards interactions in microbial communities (still related to antibiotics though). Exciting times ahead!
Note
* For some reason, the university administration refuses to call this position assistant professor in English at this time, instead referring to the position as “Postdoctoral research fellow”. I guess that it might be bloody annoying explaining that this is not the same as “postdoctoral researcher” and virtually everywhere else would be called “(non-tenured) assistant professor”, but then on the other hand, who cares about titles anyway?
New year – new challenges
So 2017 has begun, and this year will bring new challenges and exciting opportunities. First of all, my application for a 3.5 year grant from the Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning (FORMAS) to go to Prof. Jo Handelsman‘s lab in the US was granted. Since Prof. Handelsman in is moving her lab to University of Wisconsin in Madison, where she will be heading the Wisconsin Institute of Discovery (after returning from the White House), it means that this summer I will be moving to Wisconsin. I will retain a link to the University of Gothenburg and the Joakim Larsson lab though, and part of the grant is actually for covering my salary after returning from the US, so Gothenburg won’t get rid of me so easily.
The granted project will use high-throughput sequencing techniques to identify genes improving colonization and invasion ability or resistance to invasion in microbial communities, using a model system developed by the Handelsman lab. The project will focus on genes important for colonization, invasion and resistance to invasion under exposure to sub-lethal antibiotics concentrations. The project will contribute important knowledge towards the understanding of microbial colonization and invasion and highlight disturbances to the interactions in microbial communities caused by anthropogenic activities. In addition, the results of the project will hopefully allow for prediction of secondary effects of antibiotic exposure in the environment, and the preparation for future challenges related to infections with pathogenic bacteria. The project has already been highlighted by CARe (although this was before Jo announced her move from Yale) and a FORMAS press release (in Swedish).
The project will go under the acronym InSiDER, and I intend to write about it in a special section of the website, called the Wisconsin Blog. My intention is to include personal reflections on life in Wisconsin and work in the Handelsman lab there, but we’ll see how those plans turn out. Anyway, I am very thankful for FORMAS funding this project and giving me the opportunity to work with one of the leading scientists within microbial ecology in the world!