Published book chapter: Strategies for metagenomic analysis
Last summer, I was approached by Muniyandi Nagarajan to write a book chapter for a book on metagenomics. The book was published earlier this month, and is now available online (1). I have to admit that I have not yet read the entire book, but my own chapter deals with selecting the right tools for metagenomic analysis, and discusses different strategies to perform taxonomic classification, functional analysis, metagenomic assembly, and statistical comparisons between metagenomes (2). The chapter also considers the pros and cons of automated computational “pipelines” for analysis of metagenomic data. While I do not point to a specific set of software that obviously perform better in all situations, I do highlight some analysis strategies that clearly should be avoided. The chapter also suggests a few among the set of robust and well-functioning software tools that, in my opinion, should be used for metagenomic analyses. To some degree, this paper overlaps with the review paper we wrote on using metagenomics to analyze antibiotic resistance genes in various environments, published earlier this year (3), but the discussion in the book chapter is far more general. I imagine that the book chapter could be used, for example, in teaching metagenomics to students in bioinformatics (that’s at least a use I envision myself). Finally, apart from my own chapter, I can also highly recommend the chapter by Boulund et al. on statistical considerations for metagenomic data analysis (4). The book is available to buy from here, and the chapter can be read here.
References
- Nagarajan M (Ed.) Metagenomics: Perspectives, Methods, and Applications. ISBN: 9780081022689. Academic Press, Elsevier, USA (2018). doi: 10.1016/B978-0-08-102268-9 [Link]
- Bengtsson-Palme J: Strategies for Taxonomic and Functional Annotation of Metagenomes. In: Nagarajan M (Ed.) Metagenomics: Perspectives, Methods, and Applications, 55–79. Academic Press, Elsevier, USA (2018). doi: 10.1016/B978-0-08-102268-9.00003-3 [Link]
- Bengtsson-Palme J, Larsson DGJ, Kristiansson E: Using metagenomics to investigate human and environmental resistomes. Journal of Antimicrobial Chemotherapy, 72, 2690–2703 (2017). doi: 10.1093/jac/dkx199 [Paper link]
- Boulund F, Pereira MB, Jonsson V, Kristiansson E: Computational and Statistical Considerations in the Analysis of Metagenomic Data. In: Nagarajan M (Ed.) Metagenomics: Perspectives, Methods, and Applications, 81–102. Academic Press,, Elsevier, USA (2018). doi: 10.1016/B978-0-08-102268-9.00004-5 [Link]
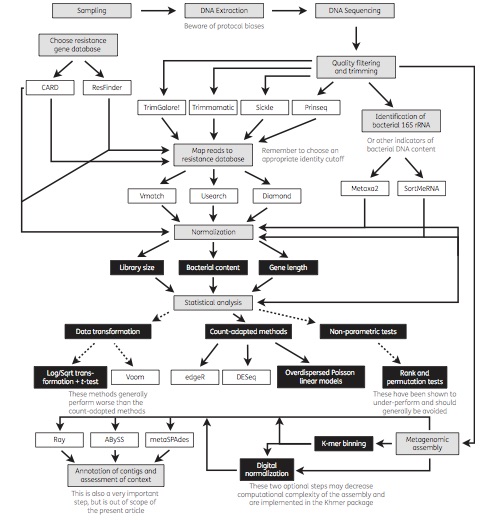
Published paper: Investigating resistomes using metagenomics
Today, a review paper which I wrote together with Joakim Larsson and Erik Kristiansson was published in Journal of Antimicrobial Chemotherapy (1). We have for a long time used metagenomic DNA sequencing to study antibiotic resistance in different environments (2-6), including in the human microbiota (7). Generally, our ultimate purpose has been to assess the risks to human health associated with resistance genes in the environment. However, a multitude of methods exist for metagenomic data analysis, and over the years we have learned that not all methods are suitable for the investigation of resistance genes for this purpose. In our review paper, we describe and discuss current methods for sequence handling, mapping to databases of resistance genes, statistical analysis and metagenomic assembly. We also provide an overview of important considerations related to the analysis of resistance genes, and end by recommending some of the currently used tools, databases and methods that are best equipped to inform research and clinical practice related to antibiotic resistance (see the figure from the paper below). We hope that the paper will be useful to researchers and clinicians interested in using metagenomic sequencing to better understand the resistance genes present in environmental and human-associated microbial communities.
References
- Bengtsson-Palme J, Larsson DGJ, Kristiansson E: Using metagenomics to investigate human and environmental resistomes. Journal of Antimicrobial Chemotherapy, advance access (2017). doi: 10.1093/jac/dkx199 [Paper link]
- Bengtsson-Palme J, Boulund F, Fick J, Kristiansson E, Larsson DGJ: Shotgun metagenomics reveals a wide array of antibiotic resistance genes and mobile elements in a polluted lake in India. Frontiers in Microbiology, 5, 648 (2014). doi: 10.3389/fmicb.2014.00648 [Paper link]
- Lundström S, Östman M, Bengtsson-Palme J, Rutgersson C, Thoudal M, Sircar T, Blanck H, Eriksson KM, Tysklind M, Flach C-F, Larsson DGJ: Minimal selective concentrations of tetracycline in complex aquatic bacterial biofilms. Science of the Total Environment, 553, 587–595 (2016). doi: 10.1016/j.scitotenv.2016.02.103 [Paper link]
- Bengtsson-Palme J, Hammarén R, Pal C, Östman M, Björlenius B, Flach C-F, Kristiansson E, Fick J, Tysklind M, Larsson DGJ: Elucidating selection processes for antibiotic resistance in sewage treatment plants using metagenomics. Science of the Total Environment, 572, 697–712 (2016). doi: 10.1016/j.scitotenv.2016.06.228 [Paper link]
- Pal C, Bengtsson-Palme J, Kristiansson E, Larsson DGJ: The structure and diversity of human, animal and environmental resistomes. Microbiome, 4, 54 (2016). doi: 10.1186/s40168-016-0199-5 [Paper link]
- Flach C-F, Pal C, Svensson CJ, Kristiansson E, Östman M, Bengtsson-Palme J, Tysklind M, Larsson DGJ: Does antifouling paint select for antibiotic resistance? Science of the Total Environment, 590–591, 461–468 (2017). doi: 10.1016/j.scitotenv.2017.01.213 [Paper link]
- Bengtsson-Palme J, Angelin M, Huss M, Kjellqvist S, Kristiansson E, Palmgren H, Larsson DGJ, Johansson A: The human gut microbiome as a transporter of antibiotic resistance genes between continents. Antimicrobial Agents and Chemotherapy, 59, 10, 6551–6560 (2015). doi: 10.1128/AAC.00933-15 [Paper link]
Published paper: FARAO
Late last year, we introduced FARAO – the Flexible All-Round Annotation Organizer – a software tool that allows visualization of annotated features on contigs. Today, the Applications Note describing the software was published as an advance access paper in Bioinformatics (1). As I have described before, storing and visualizing annotation and coverage information in FARAO has a number of advantages. FARAO is able to:
- Integrate annotation and coverage information for the same sequence set, enabling coverage estimates of annotated features
- Scale across millions of sequences and annotated features
- Filter sequences, such that only entries with annotations satisfying certain given criteria will be outputted
- Handle annotation and coverage data produced by a range of different bioinformatics tools
- Handle custom parsers through a flexible interface, allowing for adaption of the software to virtually any bioinformatic tool not supported out of the box
- Produce high-quality EPS output
- Integrate with MySQL databases
I have previously used FARAO to produce annotation figures in our paper on a polluted Indian lake (2), as well as in a paper on sewage treatment plants (which is in press and should be coming out any day now). We hope that the tool will find many more uses in other projects in the future!
References
- Hammarén R, Pal C, Bengtsson-Palme J: FARAO: The Flexible All-Round Annotation Organizer. Bioinformatics, advance access (2016). doi: 10.1093/bioinformatics/btw499 [Paper link]
- Bengtsson-Palme J, Boulund F, Fick J, Kristiansson E, Larsson DGJ: Shotgun metagenomics reveals a wide array of antibiotic resistance genes and mobile elements in a polluted lake in India. Frontiers in Microbiology, 5, 648 (2014). doi: 10.3389/fmicb.2014.00648 [Paper link]
TriMetAss updated to version 1.2
TriMetAss has today been updated to version 1.2. The new version addresses a number of minor issues, some of which I thought was fixed with the previous version. The update can be found here.
The main problem with the previous version of TriMetAss was that the Trinity developers had changed many options in the Trinity software, which rendered more recent versions of Trinity incompatible with TriMetAss. TriMetAss was not the only external software using Trinity that was affected by these changes. As far as my testing goes, these incompatibilities should now be fixed, by improved Trinity version determination in TriMetAss. This is still not a guarantee for future changes though, so just to make sure, use one of the Trinity versions tested with TriMetAss (versions v2.1.1 or trinityrnaseq_r2013_08_14).
This time I would like to thank Artemis Louyakis at the Univesity of Florida and Tatsuya Unno at the Jeju National University (Korea) for their input on TriMetAss.
FARAO – The Flexible All-Round Annotation Organizer
A problem with annotating contigs from genomic and metagenomic projects is that there are few tools that allow the visualization of the annotated features, particularly if those features come from different sources. To alleviate this problem, I have (with assistance from Rickard Hammarén and Chandan Pal) over the last years developed a new annotation and read coverage visualization package – FARAO – which we today introduce to the public. FARAO has been used to produce the basis for the the contig annotation figures in my paper on the polluted Indian lake. Storing and visualizing annotation and coverage information in FARAO has a number of advantages. FARAO is able to:
- Integrate annotation and coverage information for the same sequence set, enabling coverage estimates of annotated features
- Scale across millions of sequences and annotated features
- Filter sequences, such that only entries with annotations satisfying certain given criteria will be outputted
- Handle annotation and coverage data produced by a range of different bioinformatics tools
- Handle custom parsers through a flexible interface, allowing for adaption of the software to virtually any bioinformatic tool
- Produce high-quality EPS output
- Integrate with MySQL databases
FARAO is today moved from a private pre-release state to a public beta state. It is still possible that this version contains bug that we have not discovered in our testing. Please send me an e-mail and make us aware of the potential shortcomings of our software if you find any unexpected behavior in this version of FARAO.
TriMetAss 1.1
TriMetAss has been updated to version 1.1. The new version addresses a number of minor issues and brings two new handy features. The update can be found here.
New features:
- Multiple input files can now be specified by adding several -1 and -2 options.
- TriMetAss now automatically stops if the candidate reads are the same for two iterations in a row.
Fixed issues:
- Support for recent versions of Trinity that no longer contain the Trinity.pl script.
- A minor bug causing TriMetAss to use more memory than necessary has been fixed.
- Fixed the
--stop_totaloption so that TriMetAss actually uses this option (rather than--stop_length) - Allowed complicated paths to be supplied for the output directory.
I would like to thank users Rickard Hammarén, Dr. Tatsuya Unno, Dr. Gisle Vestergaard and Dr. Joseph Nesme for providing me with the underlying information to provide these fixes. Thanks a lot!
TriMetAss – A Trinity-based targeted metagenomics assembler
With the publication of my latest paper last week (1), I also would like to highlight some of the software underpinning the findings a bit. To get around the problem that extremely common resistance genes could be present in multiple contexts and variants, causing assembler such as Velvet (2) to perform sub-optimally, we have written a software tool that utilizes Vmatch (3) and Trinity (4) to iteratively construct contigs from reads associated with resistance genes. This could of course be used in many other situations as well, when you want to specifically assemble a certain portion of a metagenome, but suspect that that portion might be found in multiple contexts.
TriMetAss is a Perl program, employing Vmatch and Trinity to construct multi-context contigs. TriMetAss uses extracted reads associated with, e.g., resistance genes as seeds for a Vmatch search against the complete set of read pairs, extracting reads matching with at least 49 bp (by default) to any of the seed reads. These reads are then assembled using Trinity. The resulting contigs are then used as seeds for another search using Vmatch to the complete set of reads, as above. All matches (including the previously matching read pairs) are again then used for a Trinity assembly. This iterative process is repeated until a stop criteria is met, e.g. when the total number of assembled nucleotides starts to drop rather than increase. The software can be downloaded here.
References:
- Bengtsson-Palme J, Boulund F, Fick J, Kristiansson E, Larsson DGJ: Shotgun metagenomics reveals a wide array of antibiotic resistance genes and mobile elements in a polluted lake in India. Frontiers in Microbiology, 5, 648 (2014). doi: 10.3389/fmicb.2014.00648
- Zerbino DR, Birney E: Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18, 821–829 (2008). doi:10.1101/gr.074492.107
- Kurtz S: The Vmatch large scale sequence analysis software (2010). http://vmatch.de/
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al.: Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29, 644–652 (2011). doi:10.1038/nbt.1883