FEMS Microbiology Reviews Award
We have been awarded with the first best article award from FEMS Microbiology Reviews for our 2018 review Environmental factors influencing the development and spread of antibiotic resistance. I and my co-authors Joakim Larsson and Erik Kristiansson are honoured and – of course – very happy with this recognition of our work. I was interviewed in relation to the prize, an interview that can be read here. But, also, the paper is open access, so you can go and check it all out in its full glory right now!
Published paper: benchmarking resistance gene identification
Since F1000Research uses a somewhat different publication scheme than most journals, I still haven’t understood if this paper is formally published after peer review, but I start to assume it is. There have been very little changes since the last version, so hence I will be lazy and basically repost what I wrote in April when the first version (the “preprint”) was posted online. The paper (1) is the result of a workshop arranged by the JRC in Italy in 2017. It describes various challenges arising from the process of designing a benchmark strategy for bioinformatics pipelines in the identification of antimicrobial resistance genes in next generation sequencing data.
The paper discusses issues about the benchmarking datasets used, testing samples, evaluation criteria for the performance of different tools, and how the benchmarking dataset should be created and distributed. Specially, we address the following questions:
- How should a benchmark strategy handle the current and expanding universe of NGS platforms?
- What should be the quality profile (in terms of read length, error rate, etc.) of in silico reference materials?
- Should different sets of reference materials be produced for each platform? In that case, how to ensure no bias is introduced in the process?
- Should in silico reference material be composed of the output of real experiments, or simulated read sets? If a combination is used, what is the optimal ratio?
- How is it possible to ensure that the simulated output has been simulated “correctly”?
- For real experiment datasets, how to avoid the presence of sensitive information?
- Regarding the quality metrics in the benchmark datasets (e.g. error rate, read quality), should these values be fixed for all datasets, or fall within specific ranges? How wide can/should these ranges be?
- How should the benchmark manage the different mechanisms by which bacteria acquire resistance?
- What is the set of resistance genes/mechanisms that need to be included in the benchmark? How should this set be agreed upon?
- Should datasets representing different sample types (e.g. isolated clones, environmental samples) be included in the same benchmark?
- Is a correct representation of different bacterial species (host genomes) important?
- How can the “true” value of the samples, against which the pipelines will be evaluated, be guaranteed?
- What is needed to demonstrate that the original sample has been correctly characterised, in case real experiments are used?
- How should the target performance thresholds (e.g. specificity, sensitivity, accuracy) for the benchmark suite be set?
- What is the impact of these performance thresholds on the required size of the sample set?
- How can the benchmark stay relevant when new resistance mechanisms are regularly characterized?
- How is the continued quality of the benchmark dataset ensured?
- Who should generate the benchmark resource?
- How can the benchmark resource be efficiently shared?
Of course, we have not answered all these questions, but I think we have come down to a decent description of the problems, which we see as an important foundation for solving these issues and implementing the benchmarking standard. Some of these issues were tackled in our review paper from last year on using metagenomics to study resistance genes in microbial communities (2). The paper also somewhat connects to the database curation paper we published in 2016 (3), although this time the strategies deal with the testing datasets rather than the actual databases. The paper is the first outcome of the workshop arranged by the JRC on “Next-generation sequencing technologies and antimicrobial resistance” held October 4-5 2017 in Ispra, Italy. You can find the paper here (it’s open access).
On another note, the new paper describing the UNITE database (4) has now got a formal issue assigned to it, as has the paper on tandem repeat barcoding in fungi published in Molecular Ecology Resources last year (5).
References and notes
- Angers-Loustau A, Petrillo M, Bengtsson-Palme J, Berendonk T, Blais B, Chan KG, Coque TM, Hammer P, Heß S, Kagkli DM, Krumbiegel C, Lanza VF, Madec J-Y, Naas T, O’Grady J, Paracchini V, Rossen JWA, Ruppé E, Vamathevan J, Venturi V, Van den Eede G: The challenges of designing a benchmark strategy for bioinformatics pipelines in the identification of antimicrobial resistance determinants using next generation sequencing technologies. F1000Research, 7, 459 (2018). doi: 10.12688/f1000research.14509.1
- Bengtsson-Palme J, Larsson DGJ, Kristiansson E: Using metagenomics to investigate human and environmental resistomes. Journal of Antimicrobial Chemotherapy, 72, 2690–2703 (2017). doi: 10.1093/jac/dkx199
- Bengtsson-Palme J, Boulund F, Edström R, Feizi A, Johnning A, Jonsson VA, Karlsson FH, Pal C, Pereira MB, Rehammar A, Sánchez J, Sanli K, Thorell K: Strategies to improve usability and preserve accuracy in biological sequence databases. Proteomics, 16, 18, 2454–2460 (2016). doi: 10.1002/pmic.201600034
- Nilsson RH, Larsson K-H, Taylor AFS, Bengtsson-Palme J, Jeppesen TS, Schigel D, Kennedy P, Picard K, Glöckner FO, Tedersoo L, Saar I, Kõljalg U, Abarenkov K: The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Research, 47, D1, D259–D264 (2019). doi: 10.1093/nar/gky1022
- Wurzbacher C, Larsson E, Bengtsson-Palme J, Van den Wyngaert S, Svantesson S, Kristiansson E, Kagami M, Nilsson RH: Introducing ribosomal tandem repeat barcoding for fungi. Molecular Ecology Resources, 19, 1, 118–127 (2019). doi: 10.1111/1755-0998.12944
Published paper: Diarrhea-causing bacteria in the Choqueyapu River in Bolivia
My first original paper of the year was just published in PLoS ONE. This is a collaboration with Åsa Sjöling’s group at the Karolinska Institute and the Universidad Mayor de San Andrés in Bolivia, and the project has been largely run by Jessica Guzman-Otazo.
Poor drinking water quality is a major cause of diarrhea, especially in the absence of well-working sewage treatment systems. In the study, we investigate the numbers of bacteria causing diarrhea (or actually, marker genes for those bacteria) in water, soil and vegetable samples from the Choqueyapu River area in La Paz – Bolivia’s third largest city (1). The river receives sewage and wastewater from industries and hospitals while flowing through La Paz. We found that levels of ETEC – a bacterium that causes severe diarrhea – were much higher in the city than upstream of it, including at a site where the river water is used for irrigation of crops.
In addition, several multi-resistant bacteria could be isolated from the samples, of which many were emerging, globally spreading, multi-resistant variants. The results of the study indicate that there is a real risk for spreading of diarrheal diseases by using the contaminated water for drinking and irrigation (2,3). Furthermore, the identification of multi-resistant bacteria that can cause human diseases show that water contamination is an important route through which antibiotic resistance can be transferred from the environment back to humans (4).
The study was published in PLoS ONE and can be found here.
References
- Guzman-Otazo J, Gonzales-Siles L, Poma V, Bengtsson-Palme J, Thorell K, Flach C-F, Iñiguez V, Sjöling Å: Diarrheal bacterial pathogens and multi-resistant enterobacteria in the Choqueyapu River in La Paz, Bolivia. PLoS ONE, 14, 1, e0210735 (2019). doi: 10.1371/journal.pone.0210735
- Graham DW, Collignon P, Davies J, Larsson DGJ, Snape J: Underappreciated Role of Regionally Poor Water Quality on Globally Increasing Antibiotic Resistance. Environ Sci Technol 141001154428000 (2014). doi: 10.1021/es504206x
- Bengtsson-Palme J: Antibiotic resistance in the food supply chain: Where can sequencing and metagenomics aid risk assessment? Current Opinion in Food Science, 14, 66–71 (2017). doi: 10.1016/j.cofs.2017.01.010
- Bengtsson-Palme J, Kristiansson E, Larsson DGJ: Environmental factors influencing the development and spread of antibiotic resistance. FEMS Microbiology Reviews, 42, 1, 68–80 (2018). doi: 10.1093/femsre/fux053
Published paper: The UNITE database
In the 2019 database issue, Nucleic Acids Research will include a new paper on the UNITE database for molecular identification of fungi (1). I have been involved in the development of UNITE in different ways since 2012, most prominently via the ITSx (2) and Atosh software which are ticking under the hood of the database.
In this update paper, we introduce a redesigned handling of unclassifiable species hypotheses, integration with the taxonomic backbone of the Global Biodiversity Information Facility, and support for an unlimited number of parallel taxonomic classification systems. The database now contains around one million fungal ITS sequences that can be used for reference, which are clustered into roughly 459,000 species hypotheses (3). Each species hypothesis is assigned a digital object identifier (DOI), which enables unambiguous reference across studies. The paper is available as open access and the UNITE database is available open source from here.
References
- Nilsson RH, Larsson K-H, Taylor AFS, Bengtsson-Palme J, Jeppesen TS, Schigel D, Kennedy P, Picard K, Glöckner FO, Tedersoo L, Saar I, Kõljalg U, Abarenkov K: The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Research, Advance article, gky1022 (2018). doi: 10.1093/nar/gky1022
- Bengtsson-Palme J, Ryberg M, Hartmann M, Branco S, Wang Z, Godhe A, De Wit P, Sánchez-García M, Ebersberger I, de Souza F, Amend AS, Jumpponen A, Unterseher M, Kristiansson E, Abarenkov K, Bertrand YJK, Sanli K, Eriksson KM, Vik U, Veldre V, Nilsson RH: Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for use in environmental sequencing. Methods in Ecology and Evolution, 4, 10, 914–919 (2013). doi: 10.1111/2041-210X.12073
- Kõljalg U, Nilsson RH, Abarenkov K, Tedersoo L, Taylor AFS, Bahram M, Bates ST, Bruns TT, Bengtsson-Palme J, Callaghan TM, Douglas B, Drenkhan T, Eberhardt U, Dueñas M, Grebenc T, Griffith GW, Hartmann M, Kirk PM, Kohout P, Larsson E, Lindahl BD, Lücking R, Martín MP, Matheny PB, Nguyen NH, Niskanen T, Oja J, Peay KG, Peintner U, Peterson M, Põldmaa K, Saag L, Saar I, Schüßler A, Senés C, Smith ME, Suija A, Taylor DE, Telleria MT, Weiß M, Larsson KH: Towards a unified paradigm for sequence-based identification of Fungi. Molecular Ecology, 22, 21, 5271–5277 (2013). doi: 10.1111/mec.12481
Published paper: Breast milk and the infant gut resistome
This week, a paper by my former roommate Katariina Pärnänen was published by Nature Communications. In the paper (1), we use shotgun metagenomics to show that infants carry more resistant bacteria in their gut than adults do, irrespective of whether they themselves have been treated with antibiotics or not. We also found that the antibiotic resistance gene and mobile genetic element profiles of infant feces are more similar to those of their own mothers than to those of unrelated mothers. This is suggestive of a pathway of transmission of resistance genes from the mothers, and importantly we find that the mobile genetic elements in breastmilk are shared with those of the infant feces, despite vast differences in their microbiota composition. Finally, we find that termination of breastfeeding and intrapartum antibiotic prophylaxis of mothers are associated with higher abundances of specific ARGs in the infant gut. Our results suggest that infants inherit the legacy of past antibiotic consumption of their mothers via transmission of genes, but that the taxonomic composition of the microbiota still strongly dictates the overall load of resistance genes.
I am not going to dwell in to details of the study here, but I instead encourage you to read the paper (hey, it’s open access!) or the excellent popular summary that Katariina has already written. Finally, I want to emphasize the great work Katariina has put into this (I would know, since I shared room with her) and congratulate her on her own little infant!
Reference
- Pärnänen K, Karkman A, Hultman J, Lyra C, Bengtsson-Palme J, Larsson DGJ, Rautava S, Isolauri E, Salminen S, Kumar H, Satokari R, Virta M: Maternal gut and breast milk microbiota affect infant gut antibiotic resistome and mobile genetic elements. Nature Communications, 9, 3891 (2018). doi: 10.1038/s41467-018-06393-w [Paper link]
Published paper: Ribosomal tandem repeat barcoding for fungi
On Friday, Molecular Ecology Resources put online Christian Wurzbacher‘s latest paper, of which I am also a coauthor. The paper presents three sets of general primers that allow for amplification of the complete ribosomal operon from the ribosomal tandem repeats, covering all the ribosomal markers (ETS, SSU, ITS1, 5.8S, ITS2, LSU, and IGS) (1). This paper is important because it introduces a technique to utilize third generation sequencing (PacBio and Nanopore) to generate high‐quality reference data (equivalent or better than Sanger sequencing) in a high‐throughput manner. The paper shows that the quality of the Nanopore generated sequences was 99.85%, which is comparable with the 99.78% accuracy described for Sanger sequencing.
My main contribution to this paper is the consensus sequence generation script – Consension – which is available from my software page. Importantly, there are huge gaps in the reference databases we use for taxonomic classification and this method will facilitate the integration of reference data from all of the ribosomal markers. We hope that this work will stimulate large-scale generation of ribosomal reference data covering several marker genes, linking previously spread-out information together.
Reference
- Wurzbacher C, Larsson E, Bengtsson-Palme J, Van den Wyngaert S, Svantesson S, Kristiansson E, Kagami M, Nilsson RH: Introducing ribosomal tandem repeat barcoding for fungi. Molecular Ecology Resources, Accepted article (2018). doi: 10.1111/1755-0998.12944 [Paper link]
Published paper: The global topsoil microbiome
I’m really late at this ball for a number of reasons, but last week Nature published our paper on the structure and function of the global topsoil microbiome (1). This paper has a long story, but in short I got contacted by Mohammad Bahram (the first author) about two years ago about a project using metagenomic sequencing to look at a lot of soil samples for patterns of antibiotic resistance gene abundances and diversity. The project had made the interesting discovery that resistance gene abundances were linked to the ratio of fungi and bacteria (so that more fungi was linked to more resistance genes). During the following year, we together worked on deciphering these discoveries, which are now published in Nature. The paper also deals with the taxonomic patterns linked to geography (1), but as evident from the above, my main contribution here has been on the antibiotic resistance side.
In short, we find that:
- Bacterial diversity is highest in temperate habitats, and lower both closer to the equator and the poles
- For bacteria, the diversity of biological functions follows the same pattern, but for fungi, the functional diversity is higher closer to the poles and the equator
- Higher abundance of fungi is linked to higher abundance and diversity of antibiotic resistance genes. Specifically, this is related to known antibiotic producing fungal lineages, such as Penicillium and Oidiodendron. There also seems to be a link between the Actinobacteria, encompassing the antibiotic-producing bacterial genus of Streptomyces and higher resistance gene diversity.
- Similar relationships between the fungus-like Oomycetes and resistance genes was also found in ocean samples from the Tara Oceans project (2)
The results of this study indicate that both environmental filtering and niche differentiation determine soil microbial composition, and that the role of dispersal limitation is minor at this scale. Soil pH and precipitation seems to be the strongest drivers of community composition. Furthermore, we interpret our data to reveal that inter-kingdom antagonism is important in structuring microbial communities. This speaks against the notion put forward that antibiotic resistance genes might not have a resistance function in natural settings (3). That said, the most likely explanation here is probably a bit of both warfare and repurposing of genes. Soil seems to be the largest untapped source of resistance genes for human pathogens (4), and the finding that natural antagonism may be driving resistance gene diversification and enrichment may be important for future management of environmental antibiotic resistance (5,6).
It was really great to work with Mohammad and his team, and I sure hope that we will collaborate again in the future. The entire paper can be found in the issue of Nature coming out this week, and is already online at Nature’s website.
References
- Bahram M°, Hildebrand F°, Forslund SK, Anderson JL, Soudzilovskaia NA, Bodegom PM, Bengtsson-Palme J, Anslan S, Coelho LP, Harend H, Huerta-Cepas J, Medema MH, Maltz MR, Mundra S, Olsson PA, Pent M, Põlme S, Sunagawa S, Ryberg M, Tedersoo L, Bork P: Structure and function of the global topsoil microbiome. Nature, 560, 233–237 (2018). doi: 10.1038/s41586-018-0386-6
- Sunagawa S et al. Structure and function of the global ocean microbiome. Science 348, 6237, 1261359 (2015). doi: 10.1126/science.1261359
- Aminov RI: The role of antibiotics and antibiotic resistance in nature. Environmental Microbiology, 11, 12, 2970-2988 (2009). doi: 10.1111/j.1462-2920.2009.01972.x
- Bengtsson-Palme J: The diversity of uncharacterized antibiotic resistance genes can be predicted from known gene variants – but not always. Microbiome, 6, 125 (2018). doi: 10.1186/s40168-018-0508-2
- Bengtsson-Palme J, Kristiansson E, Larsson DGJ: Environmental factors influencing the development and spread of antibiotic resistance. FEMS Microbiology Reviews, 42, 1, 68–80 (2018). doi: 10.1093/femsre/fux053
- Larsson DGJ, Andremont A, Bengtsson-Palme J, Brandt KK, de Roda Husman AM, Fagerstedt P, Fick J, Flach C-F, Gaze WH, Kuroda M, Kvint K, Laxminarayan R, Manaia CM, Nielsen KM, Ploy M-C, Segovia C, Simonet P, Smalla K, Snape J, Topp E, van Hengel A, Verner-Jeffreys DW, Virta MPJ, Wellington EM, Wernersson A-S: Critical knowledge gaps and research needs related to the environmental dimensions of antibiotic resistance. Environment International, 117, 132–138 (2018).
Published paper: Predicting the uncharacterized resistome
Over the weekend, Microbiome put online my most recent paper (1) – a project which started as an idea I got when I finished up my PhD thesis in 2016. One of my main points in the thesis (2), which was also made again on our recent review on environmental factors influencing resistance development (3), is that the greatest risks associated with antibiotic resistance in the environment may not be the resistance genes already circulating in pathogens (which are relatively easily quantified), but the ones associated with recruitment of novel resistance genes from bacteria in the environment (2-4). The latter genes are, however, impossible to quantify due to the fact that they are unknown. But what if we could use knowledge of the diversity and abundance of known resistance genes to estimate the same properties of the yet uncharacterized resistome? That would be a great advantage in e.g. ranking of risk environments, as then some property that is easily monitored can be used to inform risk management of both known and unknown resistance factors.
This just published paper explores this possibility, by quantifying the abundance and diversity of resistance genes in 1109 metagenomes from various environments (1). I have taken two different approaches. First, I took out smaller subsets of genes from the reference database (in this case Resqu, a database of antibiotic resistance genes with verified resistance functions, detected on mobile genetic elements), and used those subsets to estimate resistome diversity and abundance in the 1109 metagenomes. Then these predictions were compared to the results of the entire database. I then, in a second step, investigated if these predictions could be extended to a set of truly novel resistance genes, i.e. the resistance genes present in the FARME database, collecting data from functional metagenomics inserts (5,6).
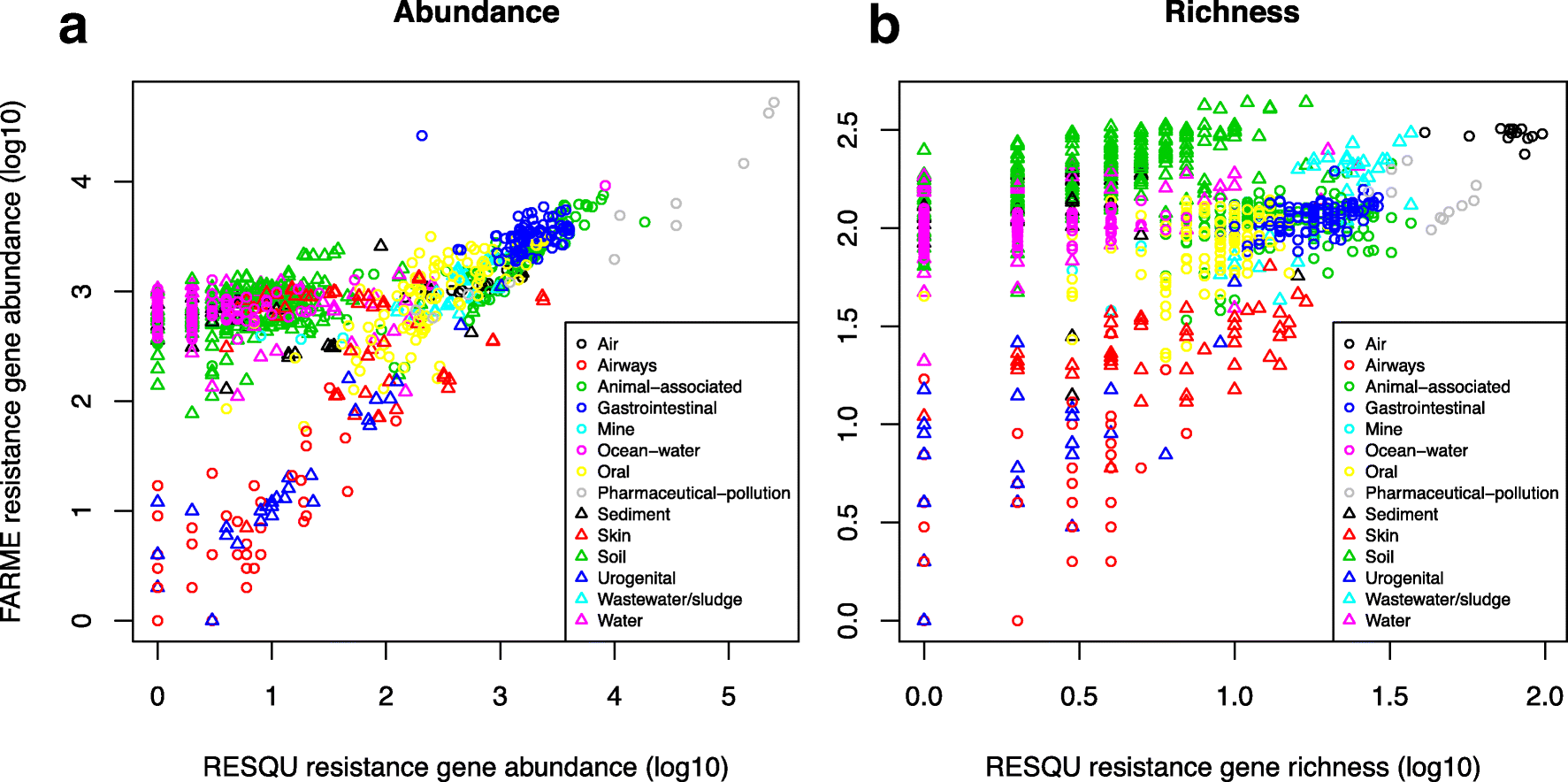
The results show that generally the diversity and abundance of known antibiotic resistance genes can be used to predict the same properties of undescribed resistance genes (see figure above). However, the extent of this predictability is, importantly, dependent on the type of environment investigated. The study also shows that carefully selected small sets of resistance genes can describe total resistance gene diversity remarkably well. This means that knowledge gained from large-scale quantifications of known resistance genes can be utilized as a proxy for unknown resistance factors. This is important for current and proposed monitoring efforts for environmental antibiotic resistance (7-11) and has implications for the design of risk ranking strategies and the choices of measures and methods for describing resistance gene abundance and diversity in the environment.
The study also investigated which diversity measures were best suited to estimate total diversity. Surprisingly, some diversity measures described the total diversity of resistance genes remarkably bad. Most prominently, the Simpson diversity index consistently showed poor performance, and while the Shannon index performed relatively better, there is still no reason to select the Shannon index over normalized (rarefied) richness of resistance genes. The ACE estimator fluctuated substantially compared to the other diversity measures, while the Chao1 estimator more consistently showed performance very similar to richness. Therefore, either richness or the Chao1 estimator should be used for ranking resistance gene diversity, while the Shannon, Simpson, and ACE measures should be avoided.
Importantly, this study implies that the recruitment of novel antibiotic resistance genes from the environment to human pathogens is essentially random. Therefore, when ranking risks associated with antibiotic resistance in environmental settings, the knowledge gained from large-scale quantification of known resistance genes can be utilized as a proxy for the unknown resistance factors (although this proxy is not perfect). Thus, high-risk environments for resistance development and dissemination would, for example, be aquaculture, animal husbandry, discharges from antibiotic manufacturing, and untreated sewage (3,8,12-15). Further attention should probably be paid to antibiotic contaminated soils, as this study points to soils as a vast source of resistance genes not yet encountered in human pathogens. This has also been suggested previously by others (16-19). The results of this study can be used to guide monitoring efforts for environmental antibiotic resistance, to design risk ranking strategies, and to choose appropriate measures and methods for describing resistance gene abundance and diversity in the environment. The entire open access paper is available here.
References
- Bengtsson-Palme J: The diversity of uncharacterized antibiotic resistance genes can be predicted from known gene variants – but not always. Microbiome, 6, 125 (2018). doi: 10.1186/s40168-018-0508-2
- Bengtsson-Palme J: Antibiotic resistance in the environment: a contribution from metagenomic studies. Doctoral thesis (medicine), Department of Infectious Diseases, Institute of Biomedicine, Sahlgrenska Academy, University of Gothenburg, 2016. [Link]
- Bengtsson-Palme J, Kristiansson E, Larsson DGJ: Environmental factors influencing the development and spread of antibiotic resistance. FEMS Microbiology Reviews, 42, 1, 68–80 (2018). doi: 10.1093/femsre/fux053
- Bengtsson-Palme J, Larsson DGJ: Antibiotic resistance genes in the environment: prioritizing risks. Nature Reviews Microbiology, 13, 369 (2015). doi: 10.1038/nrmicro3399-c1
- Wallace JC, Port JA, Smith MN, Faustian EM: FARME DB: a functional antibiotic resistance element database. Database, 2017, baw165 (2017).
- Handelsman J, Rondon MR, Brady SF, Clardy J, Goodman RM: Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chemical Biology, 5, R245–249 (1998).
- Berendonk TU, Manaia CM, Merlin C, Fatta-Kassinos D, Cytryn E, Walsh F, et al.: Tackling antibiotic resistance: the environmental framework. Nature Reviews Microbiology, 13, 310–317 (2015).
- Pruden A, Larsson DGJ, Amézquita A, Collignon P, Brandt KK, Graham DW, et al.: Management options for reducing the release of antibiotics and antibiotic resistance genes to the environment. Environmental Health Perspectives, 121, 878–885 (2013).
- Review on Antimicrobial Resistance: Antimicrobials in agriculture and the environment: reducing unnecessary use and waste. O’Neill J, ed. London: Wellcome Trust & HM Government (2015).
- Angers-Loustau A, Petrillo M, Bengtsson-Palme J, Berendonk T, Blais B, Chan KG, Coque TM, Hammer P, Heß S, Kagkli DM, Krumbiegel C, Lanza VF, Madec J-Y, Naas T, O’Grady J, Paracchini V, Rossen JWA, Ruppé E, Vamathevan J, Venturi V, Van den Eede G: The challenges of designing a benchmark strategy for bioinformatics pipelines in the identification of antimicrobial resistance determinants using next generation sequencing technologies. F1000Research, 7, 459 (2018). doi: 10.12688/f1000research.14509.1
- Larsson DGJ, Andremont A, Bengtsson-Palme J, Brandt KK, de Roda Husman AM, Fagerstedt P, Fick J, Flach C-F, Gaze WH, Kuroda M, Kvint K, Laxminarayan R, Manaia CM, Nielsen KM, Ploy M-C, Segovia C, Simonet P, Smalla K, Snape J, Topp E, van Hengel A, Verner-Jeffreys DW, Virta MPJ, Wellington EM, Wernersson A-S: Critical knowledge gaps and research needs related to the environmental dimensions of antibiotic resistance. Environment International, 117, 132–138 (2018). doi: 10.1016/j.envint.2018.04.041
- Allen HK, Donato J, Wang HH, Cloud-Hansen KA, Davies J, Handelsman J: Call of the wild: antibiotic resistance genes in natural environments. Nature Reviews Microbiology, 8, 251–259 (2010).
- Graham DW, Collignon P, Davies J, Larsson DGJ, Snape J: Underappreciated role of regionally poor water quality on globally increasing antibiotic resistance. Environmental Science & Technology, 48,11746–11747 (2014).
- Larsson DGJ: Pollution from drug manufacturing: review and perspectives. Philosophical Transactions of the Royal Society of London, Series B Biological Sciences, 369, 20130571 (2014).
- Cabello FC, Godfrey HP, Buschmann AH, Dölz HJ: Aquaculture as yet another environmental gateway to the development and globalisation of antimicrobial resistance. Lancet Infectious Diseases, 16, e127–133 (2016).
- Forsberg KJ, Reyes A, Wang B, Selleck EM, Sommer MOA, Dantas G: The shared antibiotic resistome of soil bacteria and human pathogens. Science, 337, 1107–1111 (2012).
- Allen HK, Moe LA, Rodbumrer J, Gaarder A, Handelsman J: Functional metagenomics reveals diverse beta-lactamases in a remote Alaskan soil. ISME Journal, 3, 243–251 (2009).
- Riesenfeld CS, Goodman RM, Handelsman J: Uncultured soil bacteria are a reservoir of new antibiotic resistance genes. Environmental Microbiology, 6, 981–989 (2004).
- McGarvey KM, Queitsch K, Fields S: Wide variation in antibiotic resistance proteins identified by functional metagenomic screening of a soil DNA library. Applied and Environmental Microbiology, 78, 1708–1714 (2012).
Published paper: A Metaxa2 database for the arthropod COI locus
A few days ago I posted about that Bioinformatics had published our paper on the Metaxa2 Database Builder (1). Today, I am happy to report that PeerJ has published the first paper in which the database builder is used to create a new Metaxa2 (2) database! My colleagues at Ohio State University has used the software to build a database for the COI gene (3), which is commonly used in arthropod barcoding. The used region was extracted from COI sequences from arthropod whole mitochondrion genomes, and employed to create a database containing sequences from all major arthropod clades, including all insect orders, all arthropod classes and the Onychophora, Tardigrada and Mollusca outgroups.
Similar to what we did in our evaluation of taxonomic classifiers used on non-rRNA barcoding regions (4), we performed a cross-validation analysis to characterize the relationship between the Metaxa2 reliability score, an estimate of classification confidence, and classification error probability. We used this analysis to select a reliability score threshold which minimized error. We then estimated classification sensitivity, false discovery rate and overclassification, the propensity to classify sequences from taxa not represented in the reference database.
Since the database builder was still in its early inception stages when we started doing this work, the software itself saw several improvements because of this project. We believe that our work on the COI database, as well as on the recently released database builder software, will help researchers in designing and evaluating classification databases for metabarcoding on arthropods and beyond. The database is included in the new Metaxa2 2.2 release, and is also downloadable from the Metaxa2 Database Repository (1). The open access paper can be found here.
References
- Bengtsson-Palme J, Richardson RT, Meola M, Wurzbacher C, Tremblay ED, Thorell K, Kanger K, Eriksson KM, Bilodeau GJ, Johnson RM, Hartmann M, Nilsson RH: Metaxa2 Database Builder: Enabling taxonomic identification from metagenomic and metabarcoding data using any genetic marker. Bioinformatics, advance article (2018). doi: 10.1093/bioinformatics/bty482
- Bengtsson-Palme J, Hartmann M, Eriksson KM, Pal C, Thorell K, Larsson DGJ, Nilsson RH: Metaxa2: Improved identification and taxonomic classification of small and large subunit rRNA in metagenomic data. Molecular Ecology Resources, 15, 6, 1403–1414 (2015). doi: 10.1111/1755-0998.12399
- Richardson RT, Bengtsson-Palme J, Gardiner MM, Johnson RM: A reference cytochrome c oxidase subunit I database curated for hierarchical classification of arthropod metabarcoding data. PeerJ, 6, e5126 (2018). doi: 10.7717/peerj.5126
- Richardson RT, Bengtsson-Palme J, Johnson RM: Evaluating and Optimizing the Performance of Software Commonly Used for the Taxonomic Classification of DNA Sequence Data. Molecular Ecology Resources, 17, 4, 760–769 (2017). doi: 10.1111/1755-0998.12628
Published paper: Metaxa2 Database Builder
One of the questions I have received regarding Metaxa2 is if it is possible to use it on other DNA barcodes. My answer has been “technically, yes, but it is a very cumbersome process of creating a custom database for every additional barcode”. Not anymore, the newly introduced Metaxa2 Database Builder makes this process automatic, with the user just supplying a FASTA file of sequences from the region in question and a file containing the taxonomy information for the sequences (in GenBank, NSD XML, Metaxa2 or SILVA-style formats). The preprint (1) has been out for some time, but today Bioinformatics published the paper describing the software (2).
The paper not only details how the database builder works, but also shows that it is working on a number of different barcoding regions, albeit with different results in terms of accuracy. Still, even with seemingly high misclassification rates for some DNA barcodes, the software performs better than a simple BLAST-based taxonomic assignment (76.5% vs. 41.4% correct classifications for matK, and 76.2% vs. 45.1% for tnrL). The database builder has already found use in building a COI database for anthropods (3), and we envision a range of uses in the near future.
As the paper is now published, I have also moved the Metaxa2 software (4) from beta-status to a full-worthy version 2.2 update. Hopefully, this release should be bug free, but my experience is that when the community gets their hands of the software they tend to discover things our team has missed. I would like to thank the entire team working on this, particularly Rodney Richardson (who initiated this entire thing) and Henrik Nilsson. The software can be downloaded here. Happy barcoding!
References
- Bengtsson-Palme J, Richardson RT, Meola M, Wurzbacher C, Tremblay ED, Thorell K, Kanger K, Eriksson KM, Bilodeau GJ, Johnson RM, Hartmann M, Nilsson RH: Taxonomic identification from metagenomic or metabarcoding data using any genetic marker. bioRxiv 253377 (2018). doi: 10.1101/253377 [Link]
- Bengtsson-Palme J, Richardson RT, Meola M, Wurzbacher C, Tremblay ED, Thorell K, Kanger K, Eriksson KM, Bilodeau GJ, Johnson RM, Hartmann M, Nilsson RH: Metaxa2 Database Builder: Enabling taxonomic identification from metagenomic and metabarcoding data using any genetic marker. Bioinformatics, advance article (2018). doi: 10.1093/bioinformatics/bty482 [Paper link]
- Richardson RT, Bengtsson-Palme J, Gardiner MM, Johnson RM: A reference cytochrome c oxidase subunit I database curated for hierarchical classification of arthropod metabarcoding data. PeerJ Preprints, 6, e26662v1 (2018). doi: 10.7287/peerj.preprints.26662v1 [Link]
- Bengtsson-Palme J, Hartmann M, Eriksson KM, Pal C, Thorell K, Larsson DGJ, Nilsson RH: Metaxa2: Improved identification and taxonomic classification of small and large subunit rRNA in metagenomic data. Molecular Ecology Resources, 15, 6, 1403–1414 (2015). doi: 10.1111/1755-0998.12399 [Paper link]