Published paper: Computational discovery of novel qnr genes
BMC Genomics today published a paper first-authored by my long-time colleague Fredrik Boulund, which describes a computational screen of genomes and metagenomes for novel qnr fluoroquinolone resistance genes (1). The study makes use of Fredrik’s well-designed and updated qnr-prediction pipeline, but in contrast to his previous publication based on the pipeline from 2012 (2), we here study a 20-fold larger dataset of almost 13 terabases of sequence data. Based on this data, the pipeline predicted 611 putative qnr genes, including all previously described plasmid-mediated qnr gene families. 20 of the predicted genes were previously undescribed, and of these nine were selected for experimental validation. Six of those tested genes improved the survivability under ciprofloxacin exposure when expressed in Escherichia coli. The study shows that qnr genes are almost ubiquitous in environmental microbial communities. This study also lends further credibility to the hypothesis that environmental bacterial communities can act as sources of previously uncharacterized antibiotic resistance genes (3-7). The study can be read in its entirety here.
References
- Boulund F, Berglund F, Flach C-F, Bengtsson-Palme J, Marathe NP, Larsson DGJ, Kristiansson E: Computational discovery and functional validation of novel fluoroquinolone resistance genes in public metagenomic data sets. BMC Genomics, 18, 682 (2017). doi: 10.1186/s12864-017-4064-0
- Boulund F, Johnning A, Pereira MB, Larsson DGJ, Kristiansson E: A novel method to discover fluoroquinolone antibiotic resistance (qnr) genes in fragmented nucleotide sequences. BMC Genomics, 13, 695 (2012). doi: 10.1186/1471-2164-13-695
- Bengtsson-Palme J, Larsson DGJ: Antibiotic resistance genes in the environment: prioritizing risks. Nature Reviews Microbiology, 13, 369 (2015). doi: 10.1038/nrmicro3399-c1
- Allen HK, Donato J, Wang HH et al.: Call of the wild: antibiotic resistance genes in natural environments. Nature Reviews Microbiology, 8, 251–259 (2010).
- Berendonk TU, Manaia CM, Merlin C et al.: Tackling antibiotic resistance: the environmental framework. Nature Reviews Microbiology, 13, 310–317 (2015).
- Martinez JL: Bottlenecks in the transferability of antibiotic resistance from natural ecosystems to human bacterial pathogens. Frontiers in Microbiology, 2, 265 (2011).
- Finley RL, Collignon P, Larsson DGJ et al.: The scourge of antibiotic resistance: the important role of the environment. Clinical Infectious Diseases, 57, 704–710 (2013).
Published paper: The Calanus glacialis mitogenome
Mitochondrial DNA Part B today published a mitochondrial genome announcement paper (1) in which I was involved in doing the assemblies and annotating them. The paper describes the mitogenome of Calanus glacialis, a marine planktonic copepod, which is a keystone species in the Arctic Ocean. The mitogenome is 20,674 bp long, and includes 13 protein-coding genes, 2 rRNA genes and 22 tRNA genes. While this is of course note a huge paper, we believe that this new resource will be of interest in understanding the structure and dynamics of C. glacialis populations. The main work in this paper has been carried out by Marvin Choquet at Nord University in Bodø, Norway. So hats off to him for great work, thanks Marvin! The paper can be read here.
Reference
- Choquet M, Alves Monteiro HJ, Bengtsson-Palme J, Hoarau G: The complete mitochondrial genome of the copepod Calanus glacialis. Mitochondrial DNA Part B, 2, 2, 506–507 (2017). doi: 10.1080/23802359.2017.1361357 [Paper link]
Published paper: Helicobacter pylori vs. the viable gastric microbiota
I have just returned from a week of vacation in Sicily (almost without internet access), so I am a tad late to this news, but earlier this week Infection and Immunity published our paper on the Helicobacter pylori transcriptome in gastric infection (and early stages of carcinogenesis), and how that relates to the transcriptionally active microbiota in the stomach environment (1). This paper has been long in the making (an earlier version of it was included in Kaisa Thorell’s PhD thesis (2)), but some late additional analyses did substantially strengthen our confidence in the suggestions we got from the original data.
In the paper (1) we use metatranscriptomic RNAseq to investigate the composition of the viable microbial community, and at the same time study H. pylori gene expression in stomach biopsies. The biopsies were sampled from individuals with different degrees of H. pylori infection and/or pre-malignant tissue changes. We found that H. pylori completely dominates the microbiota in infected individuals, but (somewhat surprisingly) also in the majority of individuals classified as H. pylori uninfected using traditional methods. This confirms previous reports that have detected minute quantities of H. pylori also in presumably uninfected individuals (3-6), and raises the question of how large part of the human population (if any) that is truly not infected/colonized by H. pylori. The abundance of H. pylori was correlated with the abundance of Campylobacter, Deinococcus, and Sulfurospirillum. It is unclear, however, if these genera only share the same habitat preferences as Helicobacter, or if they are specifically promoted by the presence of H. pylori (or tissue changes induced by it). We also found that genes involved in pH regulation and nickel transport were highly expressed in H. pylori, regardless of disease stage. As far as we know, this study is the first to use metatranscriptomics to study the viable microbiota of the human stomach, and we think that this is a promising approach for future studies on pathogen-microbiota interactions. The paper (in unedited format) can be read here.
References
- Thorell K, Bengtsson-Palme J, Liu OH, Gonzales RVP, Nookaew I, Rabeneck L, Paszat L, Graham DY, Nielsen J, Lundin SB, Sjöling Å: In vivo analysis of the viable microbiota and Helicobacter pylori transcriptome in gastric infection and early stages of carcinogenesis. Infection and Immunity, accepted manuscript (2017). doi: 10.1128/IAI.00031-17 [Paper link]
- Thorell K: Multi-level characterization of host and pathogen in Helicobacter pylori-associated gastric carcinogenesis. Doctoral thesis, Institute of Biomedicine, Sahlgrenska Academy, University of Gothenburg (2014). [Link]
- Bik EM, Eckburg PB, Gill SR, Nelson KE, Purdom EA, Francois F, Perez-Perez G, Blaser MJ, Reman DA: Molecular analysis of the bacterial microbiota in the human stomach. PNAS, 103:732-737 (2006).
- Dicksved J, Lindberg M, Rosenquist M, Enroth H, Jansson JK, Engstrand L: Molecular characterization of the stomach microbiota in patients with gastric cancer and in controls. Journal of Medical Microbiology, 58:509-516 (2009).
- Maldonado-Contreras A, Goldfarb KC, Godoy-Vitorino F, Karaoz U, Contreras M, Blaser MJ, Brodie EL, Dominguez-Bello MG: Structure of the human gastric bacterial community in relation to Helicobacter pylori status. ISME Journal, 5:574-579 (2011).
- Li TH, Qin Y, Sham PC, Lau KS, Chu KM, Leung WK: Alterations in Gastric Microbiota After H. Pylori Eradication and in Different Histological Stages of Gastric Carcinogenesis. Scientific Reports, 7:44935 (2017).
Published paper: Investigating resistomes using metagenomics
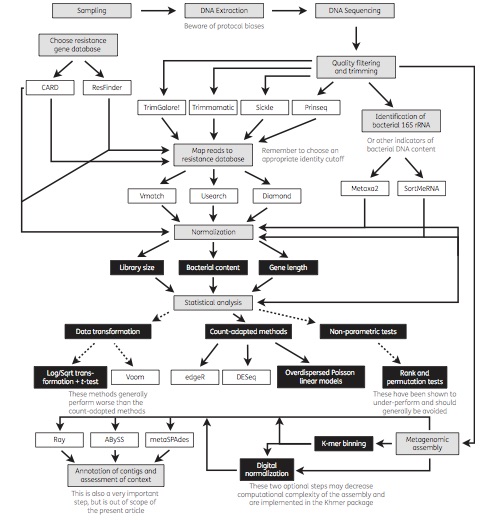
Today, a review paper which I wrote together with Joakim Larsson and Erik Kristiansson was published in Journal of Antimicrobial Chemotherapy (1). We have for a long time used metagenomic DNA sequencing to study antibiotic resistance in different environments (2-6), including in the human microbiota (7). Generally, our ultimate purpose has been to assess the risks to human health associated with resistance genes in the environment. However, a multitude of methods exist for metagenomic data analysis, and over the years we have learned that not all methods are suitable for the investigation of resistance genes for this purpose. In our review paper, we describe and discuss current methods for sequence handling, mapping to databases of resistance genes, statistical analysis and metagenomic assembly. We also provide an overview of important considerations related to the analysis of resistance genes, and end by recommending some of the currently used tools, databases and methods that are best equipped to inform research and clinical practice related to antibiotic resistance (see the figure from the paper below). We hope that the paper will be useful to researchers and clinicians interested in using metagenomic sequencing to better understand the resistance genes present in environmental and human-associated microbial communities.
References
- Bengtsson-Palme J, Larsson DGJ, Kristiansson E: Using metagenomics to investigate human and environmental resistomes. Journal of Antimicrobial Chemotherapy, advance access (2017). doi: 10.1093/jac/dkx199 [Paper link]
- Bengtsson-Palme J, Boulund F, Fick J, Kristiansson E, Larsson DGJ: Shotgun metagenomics reveals a wide array of antibiotic resistance genes and mobile elements in a polluted lake in India. Frontiers in Microbiology, 5, 648 (2014). doi: 10.3389/fmicb.2014.00648 [Paper link]
- Lundström S, Östman M, Bengtsson-Palme J, Rutgersson C, Thoudal M, Sircar T, Blanck H, Eriksson KM, Tysklind M, Flach C-F, Larsson DGJ: Minimal selective concentrations of tetracycline in complex aquatic bacterial biofilms. Science of the Total Environment, 553, 587–595 (2016). doi: 10.1016/j.scitotenv.2016.02.103 [Paper link]
- Bengtsson-Palme J, Hammarén R, Pal C, Östman M, Björlenius B, Flach C-F, Kristiansson E, Fick J, Tysklind M, Larsson DGJ: Elucidating selection processes for antibiotic resistance in sewage treatment plants using metagenomics. Science of the Total Environment, 572, 697–712 (2016). doi: 10.1016/j.scitotenv.2016.06.228 [Paper link]
- Pal C, Bengtsson-Palme J, Kristiansson E, Larsson DGJ: The structure and diversity of human, animal and environmental resistomes. Microbiome, 4, 54 (2016). doi: 10.1186/s40168-016-0199-5 [Paper link]
- Flach C-F, Pal C, Svensson CJ, Kristiansson E, Östman M, Bengtsson-Palme J, Tysklind M, Larsson DGJ: Does antifouling paint select for antibiotic resistance? Science of the Total Environment, 590–591, 461–468 (2017). doi: 10.1016/j.scitotenv.2017.01.213 [Paper link]
- Bengtsson-Palme J, Angelin M, Huss M, Kjellqvist S, Kristiansson E, Palmgren H, Larsson DGJ, Johansson A: The human gut microbiome as a transporter of antibiotic resistance genes between continents. Antimicrobial Agents and Chemotherapy, 59, 10, 6551–6560 (2015). doi: 10.1128/AAC.00933-15 [Paper link]
Published paper: Does antifouling paint select for antibiotic resistance?
After the usual (1,2) long wait between acceptance and publication, Science of the Total Environment today put a paper online in which I have played a role in the bioinformatic analysis. In the paper, we investigate whether antifouling paint containing copper and zinc could co-select for antibiotic resistance, using microbiological methods and metagenomic sequencing (3).
In this work, we have studied marine microbial biofilms allowed to grow on surfaces painted with antifouling paint submerged in sea water. Such antifouling paints often contain metals that potentially could co-select for antibiotic resistance (4). Using microbiological culturing, we found that the heavy-metal based paint co-selected for bacteria resistant to tetracycline. However, the paint did not enrich neither the total abundance of known mobile antibiotic resistance genes nor the abundance of tetracycline resistance genes in the biofilm communities. Rather, the communities from the painted surfaces were enriched for bacteria with genetic profiles suggesting increased capacity for extrusion of antibiotics via RND efflux systems. In addition, these communities were also enriched for genes involved in mobilization of DNA, such as ISCR transposases and integrases. Finally, the biofilm communities from painted surfaces displayed lower taxonomic diversity and were at the same time enriched for Gammaproteobacteria. The paper builds on our previous work in which we identify certain co-occurences between genes conferring metal and antibiotic resistance (4). However, the findings of this paper do not lend support for that mobile resistance genes are co-selected for by copper and zinc in the marine environment – rather the increase in antibiotic resistance seem to be due to taxonomic changes and cross-resistance mechanisms. The entire paper can be read here.
References
- Bengtsson-Palme J: Published paper: Community MSCs for tetracycline. https://microbiology.se/2016/03/22/published-paper-community-mscs-for-tetracycline/
- Bengtsson-Palme J: Published paper: Antibiotic resistance in sewage treatment plants . https://microbiology.se/2016/08/17/published-paper-antibiotic-resistance-in-sewage-treatment-plants/
- Flach C-F, Pal C, Svensson CJ, Kristiansson E, Östman M, Bengtsson-Palme J, Tysklind M, Larsson DGJ: Does antifouling paint select for antibiotic resistance? Science of the Total Environment, in press (2017). doi: 10.1016/j.scitotenv.2017.01.213 [Paper link]
- Pal C, Bengtsson-Palme J, Kristiansson E, Larsson DGJ: Co-occurrence of resistance genes to antibiotics, biocides and metals reveals novel insights into their co-selection potential. BMC Genomics, 16, 964 (2015). doi: 10.1186/s12864-015-2153-5 [Paper link]
Published paper: Antibiotic resistance in the food supply chain
I am happy to announce that the opinion/review piece I wrote for Current Opinion in Food Science has been published. The paper (1) extends on some of my thoughts on how high-throughput sequencing and metagenomics can aid in risk assessment of antibiotic resistant bacteria that I outlined in my PhD thesis (2), but specifically focuses on the food supply chain and its role in resistance dissemination and selection.
In the paper, I argue for that the food supply chain is a special type of setting in the resistance puzzle, as it not only serves as a connection between environmental habitats for bacteria and humans, but also sometimes presents a substantial selection for resistance, due to use of antibiotics in agri- and aquaculture. International food standards are clear that both selection and dissemination of foodborne resistance should be considered in the risk analysis of food production (3). However, the current main use of DNA sequencing in food safety is whole genome sequencing to delineate which specific strains that are involved in foodborne disease outbreaks, including the resistance factors they may carry (4,5). Further, I argue that while shotgun metagenomics could be used to screen samples for a large number of genes involved in resistance and virulence in the food supply chain, it would at present be very costly and therefore of doubtful benefit to employ in routine screening programs. Still, metagenomics can contribute knowledge that can be used in quantitative risk assessment of antibiotic resistance in the food supply chain.
The entire paper can be read here.
References
- Bengtsson-Palme J: Antibiotic resistance in the food supply chain: Where can sequencing and metagenomics aid risk assessment? Current Opinion in Food Science, in press (2017). doi: 10.1016/j.cofs.2017.01.010 [Paper link]
- Bengtsson-Palme J: Antibiotic resistance in the environment: a contribution from metagenomic studies. Doctoral thesis (medicine), Department of Infectious Diseases, Institute of Biomedicine, Sahlgrenska Academy, University of Gothenburg, 2016. [Link]
- Codex Alimentarius Commission: Guidelines for risk analysis of foodborne antimicrobial resistance. Food and Agriculture Organization of the United Nations & World Health Organization2011. [Link]
- Franz E, Gras LM, Dallman T: Significance of whole genome sequencing for surveillance, source attribution and microbial risk assessment of foodborne pathogens. Current Opinion in Food Science, 8, 74-79 (2016). doi: 10.1016/j.cofs.2016.04.004
- Stasiewicz MJ, Bakker den HC, Wiedmann M: Genomics tools in microbial food safety. Current Opinion in Food Science, 4, 105-110 (2015). doi: 10.1016/j.cofs.2015.06.002
Published paper: Evaluating taxonomic classification software
Yesterday, Molecular Ecology Resources put online an unedited version of a recent paper which I co-authored. This time, Rodney Richardson at Ohio State University has made a tremendous work of evaluating three taxonomic classification software – the RDP Naïve Bayesian Classifier, RTAX and UTAX – on a set of DNA barcoding regions commonly used for plants, namely the ITS2, and the matK, rbcL, trnL and trnH genes.
In the paper (1), we discuss the results, merits and limitations of the classifiers. In brief, we found that:
- There is a considerable trade-off between accuracy and sensitivity for the classifiers tested, which indicates a need for improved sequence classification tools (2)
- UTAX was superior with respect to error rate, but was exceedingly stringent and thus suffered from a low assignment rate
- The RDP Naïve Bayesian Classifier displayed high sensitivity and low error at the family and order levels, but had a genus-level error rate of 9.6 percent
- RTAX showed high sensitivity at all taxonomic ranks, but at the same time consistently produced the high error rates
- The choice of locus has significant effects on the classification sensitivity of all tested tools
- All classifiers showed strong relationships between database completeness, classification sensitivity and classification accuracy
We believe that the methods of comparison we have used are simple and robust, and thereby provides a methodological and conceptual foundation for future software evaluations. On a personal note, I will thoroughly enjoy working with Rodney and Reed again; I had a great time discussing the ins and outs of taxonomic classification with them! The paper can be found here.
References and notes
- Richardson RT, Bengtsson-Palme J, Johnson RM: Evaluating and Optimizing the Performance of Software Commonly Used for the Taxonomic Classification of DNA Sequence Data. Molecular Ecology Resources, Early view (2016). doi: 10.1111/1755-0998.12628 [Paper link]
- This is something that several classifiers also showed in the evaluation we did for the Metaxa2 paper (3). Interestingly enough, Metaxa2 is better at maintaining high accuracy also as sensitivity is increased.
- Bengtsson-Palme J, Hartmann M, Eriksson KM, Pal C, Thorell K, Larsson DGJ, Nilsson RH: Metaxa2: Improved identification and taxonomic classification of small and large subunit rRNA in metagenomic data. Molecular Ecology Resources, 15, 6, 1403–1414 (2015). doi: 10.1111/1755-0998.12399 [Paper link]
Published paper: The global resistome
Late yesterday, Microbiome put online our most recent work, covering the resistomes to antibiotics, biocides and metals across a vast range of environments. In the paper (1), we perform the largest characterization of resistance genes, mobile genetic elements (MGEs) and bacterial taxonomic compositions to date, covering 864 different metagenomes from humans (350), animals (145) and external environments such as soil, water, sewage, and air (369 in total). All the investigated metagenomes were sequenced to at least 10 million reads each, using Illumina technology, making the results more comparable across environments than in previous studies (2-4).
We found that the environment types had clear differences both in terms of resistance profiles and bacterial community composition. Humans and animals hosted microbial communities with limited taxonomic diversity as well as low abundance and diversity of biocide/metal resistance genes and MGEs. On the contrary, the abundance of ARGs was relatively high in humans and animals. External environments, on the other hand, showed high taxonomic diversity and high diversity of biocide/metal resistance genes and MGEs. Water, sediment and soil generally carried low relative abundance and few varieties of known ARGs, whereas wastewater and sludge were on par with the human gut. The environments with the largest relative abundance and diversity of ARGs, including genes encoding resistance to last resort antibiotics, were those subjected to industrial antibiotic pollution and air samples from a Beijing smog event.
A paper investigating this vast amount of data is of course hard to describe in a blog post, so I strongly suggest the interested reader to head over to Microbiome’s page and read the full paper (1). However, here’s a ver short summary of the findings:
- The median relative abundance of ARGs across all environments was 0.035 copies per bacterial 16S rRNA
- Antibiotic-polluted environments have (by far) the highest abundances of ARGs
- Urban air samples carried high abundance and diversity of ARGs
- Human microbiota has high abundance and diversity of known ARGs, but low taxonomic diversity compared to the external environment
- The human and animal resistomes are dominated by tetracycline resistance genes
- Over half of the ARGs were only detected in external environments, while 20.5 % were found in human, animal and at least one of the external environments
- Biocide and metal resistance genes are more common in external environments than in the human microbiota
- Human microbiota carries low abundance and richness of MGEs compared to most external environments
Importantly, less than 1.5 % of all detected ARGs were found exclusively in the human microbiome. At the same time, 57.5 % of the known ARGs were only detected in metagenomes from environmental samples, despite that the majority of the investigated ARGs were initially encountered in pathogens. Still, our analysis suggests that most of these genes are relatively rare in the human microbiota. Environmental samples generally contained a wider distribution of resistance genes to a more diverse set of antibiotics classes. For example, the relative abundance of beta-lactam resistance genes was much larger in external environments than in human and animal microbiomes. This suggests that the external environment harbours many more varieties of resistance genes than the ones currently known from the clinic. Indeed, functional metagenomics has resulted in the discovery of many novel ARGs in external environments (e.g. 5). This all fits well with an overall much higher taxonomic diversity of environmental microbial communities. In terms of consequences associated with the potential transfer of ARGs to human pathogens, we argue that unknown resistance genes are of greater concern than those already known to circulate among human-associated bacteria (6).
This study describes the potential for many external environments, including those subjected to pharmaceutical pollution, air and wastewater/sludge, to serve as hotspots for resistance development and/or transmission of ARGs. In addition, our results indicate that these environments may play important roles in the mobilization of yet unknown ARGs and their further transmission to human pathogens. To provide guidance for risk-reducing actions we – based on this study – suggest strict regulatory measures of waste discharges from pharmaceutical industries and encourage more attention to air in the transmission of antibiotic resistance (1).
References
- Pal C, Bengtsson-Palme J, Kristiansson E, Larsson DGJ: The structure and diversity of human, animal and environmental resistomes. Microbiome, 4, 54 (2016). doi: 10.1186/s40168-016-0199-5
- Durso LM, Miller DN, Wienhold BJ. Distribution and quantification of antibiotic resistant genes and bacteria across agricultural and non-agricultural metagenomes. PLoS One. 2012;7:e48325.
- Nesme J, Delmont TO, Monier J, Vogel TM. Large-scale metagenomic-based study of antibiotic resistance in the environment. Curr Biol. 2014;24:1096–100.
- Fitzpatrick D, Walsh F. Antibiotic resistance genes across a wide variety of metagenomes. FEMS Microbiol Ecol. 2016. doi:10.1093/femsec/fiv168.
- Allen HK, Moe LA, Rodbumrer J, Gaarder A, Handelsman J. Functional metagenomics reveals diverse β-lactamases in a remote Alaskan soil. ISME J. 2009;3:243–51.
- Bengtsson-Palme J, Larsson DGJ: Antibiotic resistance genes in the environment: prioritizing risks. Nature Reviews Microbiology, 13, 369 (2015). doi: 10.1038/nrmicro3399-c1
Published opinion piece: Why limit antibiotic pollution?
Me and Joakim Larsson wrote an opinion/summary piece for the APUA Newsletter, issued by the Alliance for Prudent Use of Antibiotics, that was published yesterday (1). The paper is essentially a summary of work included in my PhD thesis, and discusses how to establish minimal selective concentrations of antibiotics for microbial communities (2-4), how to identify risk environments for resistance selection (5-9), and which mitigation strategies that can be implemented (10-12). Partially, we also discussed these issues earlier in our paper in the Medicine Maker (10), but this paper goes deeper into why limiting antibiotic pollution is important to mitigate the accelerating antibiotic resistance problem. I recommend this short summary piece to anyone who would like a brief overview of our research on antibiotic resistance, and think that it can serve as a great starting point for further reading! In addition, this issue of the newsletter features very interesting pieces on reducing antibiotics use (and disposal) outside of the clinics (13) and revival of old antibiotics (14). Please go ahead to the APUA web site and read the entire newsletter!
References
- Bengtsson-Palme J, Larsson DGJ: Why limit antibiotic pollution? The role of environmental selection in antibiotic resistance development. APUA Newsletter, 34, 2, 6-9 (2016). [Paper link].
- Bengtsson-Palme J, Larsson DGJ: Concentrations of antibiotics predicted to select for resistant bacteria: Proposed limits for environmental regulation. Environment International, 86, 140-149 (2016). doi: 10.1016/j.envint.2015.10.015 [Paper link]
- Gullberg E, Cao S, Berg OG, Ilbäck C, Sandegren L, Hughes D, et al.: Selection of resistant bacteria at very low antibiotic concentrations. PLoS Pathogens 7, e1002158 (2011).
- Lundström S, Östman M, Bengtsson-Palme J, Rutgersson C, Thoudal M, Sircar T, Blanck H, Eriksson KM, Tysklind M, Flach C-F, Larsson DGJ: Minimal selective concentrations of tetracycline in complex aquatic bacterial biofilms. Science of the Total Environment, 553, 587–595 (2016). doi: 10.1016/j.scitotenv.2016.02.103
- Bengtsson-Palme J, Boulund F, Fick J, Kristiansson E, Larsson DGJ: Shotgun metagenomics reveals a wide array of antibiotic resistance genes and mobile elements in a polluted lake in India. Frontiers in Microbiology, 5, 648 (2014). doi: 10.3389/fmicb.2014.00648
- Bengtsson-Palme J, Hammarén R, Pal C, Östman M, Björlenius B, Flach C-F, Kristiansson E, Fick J, Tysklind M, Larsson DGJ: Elucidating selection processes for antibiotic resistance in sewage treatment plants using metagenomics. Science of the Total Environment, in press (2016). doi: 10.1016/j.scitotenv.2016.06.228
- Berendonk TU, Manaia CM, Merlin C, Fatta-Kassinos D, Cytryn E, Walsh F, et al.: Tackling antibiotic resistance: the environmental framework. Nature Reviews Microbiology, 13, 310–317 (2015). doi: 10.1038/nrmicro3439
- Martinez JL, Coque TM, Baquero F: What is a resistance gene? Ranking risk in resistomes. Nature Reviews Microbiology 2015, 13:116–123. doi:10.1038/nrmicro3399
- Bengtsson-Palme J, Larsson DGJ: Antibiotic resistance genes in the environment: prioritizing risks. Nature Reviews Microbiology, 13, 369 (2015) doi:10.1038/nrmicro3399‐c1
- Bengtsson-Palme J, Larsson DGJ: Time to limit antibiotic pollution. The Medicine Maker, 0416, 302, 17–18 (2016). [Paper link]
- Ashbolt NJ, Amézquita A, Backhaus T, Borriello P, Brandt KK, Collignon P, et al.: Human Health Risk Assessment (HHRA) for Environmental Development and Transfer of Antibiotic Resistance. Environmental Health Perspectives, 121, 993–1001 (2013)
- Pruden A, Larsson DGJ, Amézquita A, Collignon P, Brandt KK, Graham DW, et al.: Management options for reducing the release of antibiotics and antibiotic resistance genes to the environment. Environmental Health Perspectives, 121, 878–85 (2013).
- Theuretzbacher U: Optimizing the Use of Old Antibiotics — A Global Health Agenda. APUA Newsletter, 34, 2, 10-13 (2016). [Paper link].
- Amábile-Cuevas CF: Antibiotics and Antibiotic Resistance All Around Us. APUA Newsletter, 34, 2, 3-5 (2016). [Paper link].
Database quality paper in special issue
I just want to highlight that the paper on strategies to improve database accuracy and usability we recently published in Proteomics (1) has been included in their most recent issue, which is a special issue focusing on Data Quality Issues in Proteomics. I highly recommend reading our paper (of course) and many of the other in the special issue. Happy reading!
On another note, I will be giving a talk next Wednesday (October 5th) on a seminar day on next generation sequencing in clinical microbiology, titled “Antibiotic resistance in the clinic and the environment – There and back again“. You are very welcome to the lecture hall at floor 3 in our building at Guldhedsgatan 10A here in Gothenburg if you are interested! (Bear in mind though that it all starts at 8.15 in the morning.)
Finally, it seems that I am going to the Next Generation Sequencing Congress in London this year, which will be very fun! Hope to see some of you dealing with sequencing there!
References
- Bengtsson-Palme J, Boulund F, Edström R, Feizi A, Johnning A, Jonsson VA, Karlsson FH, Pal C, Pereira MB, Rehammar A, Sánchez J, Sanli K, Thorell K: Strategies to improve usability and preserve accuracy in biological sequence databases. Proteomics, 16, 18, 2454–2460 (2016). doi: 10.1002/pmic.201600034 [Paper link]