Major ITSx update (beta version)
Today, I am very happy to announce that after years in the making and months in testing, the next generation of ITSx, version 1.1, is ready to step into the public light and scrutiny. I have today uploaded a public beta version of the ITSx 1.1 release, which I encourage everyone that have enjoyed using ITSx to try out.
The 1.1 release of ITSx includes a wide range of new feature, including:
- A 2-10x performance increase (depending on the dataset), since ITSx now utilizes hmmsearch instead of hmmscan to detect the ITS regions and distributes the CPU cores better
- Improved ITS detection among fungi and chlorophyta, by addition of new HMM-profiles
- The HMM profile format for ITSx has been updated to HMMER3/f (thus ITSx now requires HMMER version 3.1 or later)
- Better handling of interrupted HMMER searches
- Added the
--require_anchoroption to only include sequences where the complete anchor is found in the output - Added the possibility for partial sequence output for the SSU, LSU and 5.8S regions
- Fixed a bug causing problems when reading sequence data from standard input
A lot of the code has changed in this version, which means that there might still be bugs lingering in the program. Since I will be on vacation throughout July, I encourage everyone to submit bug reports and questions, but I will not promise to respond to them until in August.
I hope that you will enjoy this new ITSx release, which you can download here. Happy barcoding!
Published paper: Investigating resistomes using metagenomics
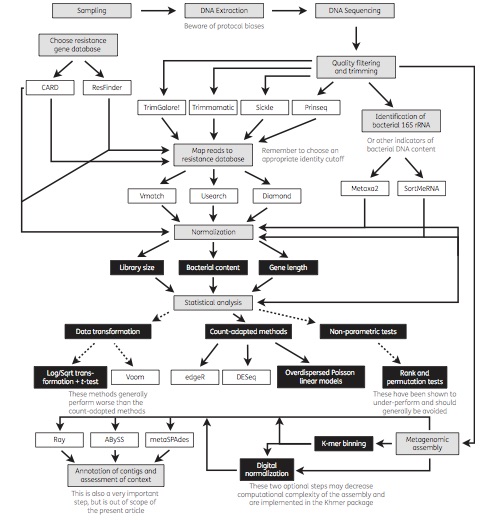
Today, a review paper which I wrote together with Joakim Larsson and Erik Kristiansson was published in Journal of Antimicrobial Chemotherapy (1). We have for a long time used metagenomic DNA sequencing to study antibiotic resistance in different environments (2-6), including in the human microbiota (7). Generally, our ultimate purpose has been to assess the risks to human health associated with resistance genes in the environment. However, a multitude of methods exist for metagenomic data analysis, and over the years we have learned that not all methods are suitable for the investigation of resistance genes for this purpose. In our review paper, we describe and discuss current methods for sequence handling, mapping to databases of resistance genes, statistical analysis and metagenomic assembly. We also provide an overview of important considerations related to the analysis of resistance genes, and end by recommending some of the currently used tools, databases and methods that are best equipped to inform research and clinical practice related to antibiotic resistance (see the figure from the paper below). We hope that the paper will be useful to researchers and clinicians interested in using metagenomic sequencing to better understand the resistance genes present in environmental and human-associated microbial communities.
References
- Bengtsson-Palme J, Larsson DGJ, Kristiansson E: Using metagenomics to investigate human and environmental resistomes. Journal of Antimicrobial Chemotherapy, advance access (2017). doi: 10.1093/jac/dkx199 [Paper link]
- Bengtsson-Palme J, Boulund F, Fick J, Kristiansson E, Larsson DGJ: Shotgun metagenomics reveals a wide array of antibiotic resistance genes and mobile elements in a polluted lake in India. Frontiers in Microbiology, 5, 648 (2014). doi: 10.3389/fmicb.2014.00648 [Paper link]
- Lundström S, Östman M, Bengtsson-Palme J, Rutgersson C, Thoudal M, Sircar T, Blanck H, Eriksson KM, Tysklind M, Flach C-F, Larsson DGJ: Minimal selective concentrations of tetracycline in complex aquatic bacterial biofilms. Science of the Total Environment, 553, 587–595 (2016). doi: 10.1016/j.scitotenv.2016.02.103 [Paper link]
- Bengtsson-Palme J, Hammarén R, Pal C, Östman M, Björlenius B, Flach C-F, Kristiansson E, Fick J, Tysklind M, Larsson DGJ: Elucidating selection processes for antibiotic resistance in sewage treatment plants using metagenomics. Science of the Total Environment, 572, 697–712 (2016). doi: 10.1016/j.scitotenv.2016.06.228 [Paper link]
- Pal C, Bengtsson-Palme J, Kristiansson E, Larsson DGJ: The structure and diversity of human, animal and environmental resistomes. Microbiome, 4, 54 (2016). doi: 10.1186/s40168-016-0199-5 [Paper link]
- Flach C-F, Pal C, Svensson CJ, Kristiansson E, Östman M, Bengtsson-Palme J, Tysklind M, Larsson DGJ: Does antifouling paint select for antibiotic resistance? Science of the Total Environment, 590–591, 461–468 (2017). doi: 10.1016/j.scitotenv.2017.01.213 [Paper link]
- Bengtsson-Palme J, Angelin M, Huss M, Kjellqvist S, Kristiansson E, Palmgren H, Larsson DGJ, Johansson A: The human gut microbiome as a transporter of antibiotic resistance genes between continents. Antimicrobial Agents and Chemotherapy, 59, 10, 6551–6560 (2015). doi: 10.1128/AAC.00933-15 [Paper link]
A note on FARAO and graphical output
I just wanted to share an experience with the FARAO software we recently published a paper about, and its compatibility with the GD and libpng libraries (used for creating PNG files). I have got questions from users about how to get this to work, and to test it out I decided to try to install it on my Mac. It turned out that it is nearly impossible to get this to work. These two packages are extremely picky with versions and dependencies. After trying for about on hour, I gave up and turned to my Linux machine. Surprisingly, I could not get it to work from scratch there either, despite that I have had it running (with some previous version combination) when we programmed and tested FARAO.
I find this extremely annoying myself, and I will try to look into other solutions for PNG or JPEG output from FARAO. In the mean time, I can only recommend to instead use the EPS output option, which produces more nice-looking figures and is considerably easier to set up. I am sorry about this and hope to be able to provide a better solution soon.
Published paper: FARAO
Late last year, we introduced FARAO – the Flexible All-Round Annotation Organizer – a software tool that allows visualization of annotated features on contigs. Today, the Applications Note describing the software was published as an advance access paper in Bioinformatics (1). As I have described before, storing and visualizing annotation and coverage information in FARAO has a number of advantages. FARAO is able to:
- Integrate annotation and coverage information for the same sequence set, enabling coverage estimates of annotated features
- Scale across millions of sequences and annotated features
- Filter sequences, such that only entries with annotations satisfying certain given criteria will be outputted
- Handle annotation and coverage data produced by a range of different bioinformatics tools
- Handle custom parsers through a flexible interface, allowing for adaption of the software to virtually any bioinformatic tool not supported out of the box
- Produce high-quality EPS output
- Integrate with MySQL databases
I have previously used FARAO to produce annotation figures in our paper on a polluted Indian lake (2), as well as in a paper on sewage treatment plants (which is in press and should be coming out any day now). We hope that the tool will find many more uses in other projects in the future!
References
- Hammarén R, Pal C, Bengtsson-Palme J: FARAO: The Flexible All-Round Annotation Organizer. Bioinformatics, advance access (2016). doi: 10.1093/bioinformatics/btw499 [Paper link]
- Bengtsson-Palme J, Boulund F, Fick J, Kristiansson E, Larsson DGJ: Shotgun metagenomics reveals a wide array of antibiotic resistance genes and mobile elements in a polluted lake in India. Frontiers in Microbiology, 5, 648 (2014). doi: 10.3389/fmicb.2014.00648 [Paper link]
Metaxa turns five years today
Today marks the five year anniversary for the Metaxa software’s initial release. Much has happened to the software since; Metaxa started off as an rRNA extraction utility for metagenomic data (1), including coarse classification to organism/organelle type. Since it has gained full-scale taxonomic classification ability better or on par with other software packages (2), much greater speed, support for the LSU gene, gained a range of related software tools (3), and spurred development of other tools such as ITSx (4). I have also been involved in no less than four peer-reviewed publications directly related to the software (1-3,5).
But it does not end here; these five years were just the beginning. We are – in different constellations – working on further enhancements to Metaxa2, including support for more genes, an updated classification database, and better customization options. I am very much still devoted to keep Metaxa2 alive and relevant as a tool for taxonomic analysis of metagenomes, applicable whenever accuracy is a key parameter. Thanks for being part of the community for these five years!
References
- Bengtsson J, Eriksson KM, Hartmann M, Wang Z, Shenoy BD, Grelet G, Abarenkov K, Petri A, Alm Rosenblad M, Nilsson RH: Metaxa: A software tool for automated detection and discrimination among ribosomal small subunit (12S/16S/18S) sequences of archaea, bacteria, eukaryotes, mitochondria, and chloroplasts in metagenomes and environmental sequencing datasets. Antonie van Leeuwenhoek, 100, 3, 471–475 (2011). doi:10.1007/s10482-011-9598-6. [Paper link]
- Bengtsson-Palme J, Hartmann M, Eriksson KM, Pal C, Thorell K, Larsson DGJ, Nilsson RH: Metaxa2: Improved identification and taxonomic classification of small and large subunit rRNA in metagenomic data. Molecular Ecology Resources, 15, 6, 1403–1414 (2015). doi: 10.1111/1755-0998.12399 [Paper link]
- Bengtsson-Palme J, Thorell K, Wurzbacher C, Sjöling Å, Nilsson RH: Metaxa2 Diversity Tools: Easing microbial community analysis with Metaxa2. Ecological Informatics, 33, 45–50 (2016). doi: 10.1016/j.ecoinf.2016.04.004 [Paper link]
- Bengtsson-Palme J, Ryberg M, Hartmann M, Branco S, Wang Z, Godhe A, De Wit P, Sánchez-García M, Ebersberger I, de Souza F, Amend AS, Jumpponen A, Unterseher M, Kristiansson E, Abarenkov K, Bertrand YJK, Sanli K, Eriksson KM, Vik U, Veldre V, Nilsson RH: Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for use in environmental sequencing. Methods in Ecology and Evolution, 4, 10, 914–919 (2013). doi: 10.1111/2041-210X.12073 [Paper link]
- Bengtsson-Palme J, Hartmann M, Eriksson KM, Nilsson RH: Metaxa, overview. In:Nelson K. (Ed.) Encyclopedia of Metagenomics: SpringerReference (www.springerreference.com). Springer-Verlag Berlin Heidelberg (2013). doi: 10.1007/978-1-4614-6418-1_239-6 [Link]
Published paper: Metaxa2 Diversity Tools
Yesterday, Ecological Informatics put our paper describing Metaxa2 Diversity Tools online (1). Metaxa2 Diversity Tools was introduced with Metaxa2 version 2.1 and consists of
- metaxa2_dc – a tool for collecting several .taxonomy.txt output files into one large abundance matrix, suitable for analysis in, e.g., R
- metaxa2_rf – generates resampling rarefaction curves (2) based on the .taxonomy.txt output
- metaxa2_si – species inference based on guessing species data from the other species present in the .taxonomy.txt output file
- metaxa2_uc – a tool for determining if the community composition of a sample is significantly different from others through resampling analysis
At the same time as I did this update to the web site, I also took the opportunity to update the Metaxa2 FAQ to better reflect recent updates to the Metaxa2 software.
Metaxa2 Diversity Tools
One often requested feature of Metaxa2 (3) has been the ability to make simple analyses from the data after classification. The Metaxa2 Diversity Tools included in Metaxa2 2.1 is a seed for such an effort (although not close to a full-fledged community analysis package comparable to QIIME (4) or Mothur (5)). It currently consist of four tools.
The Metaxa2 Data Collector (metaxa2_dc) is the simplest of them (but probably the most requested), designed to merge the output of several *.level_X.txt files from the Metaxa2 Taxonomic Traversal Tool into one large abundance matrix, suitable for further analysis in, for example, R. The Metaxa2 Species Inference tool (metaxa2_si) can be used to further infer taxon information on, for example, the species level at a lower reliability than what would be permitted by the Metaxa2 classifier, using a complementary algorithm. The idea is that is if only a single species is present in, e.g., a family and a read is assigned to this family, but not classified to the species level, that sequence will be inferred to the same species as the other reads, given that it has more than 97% sequence identity to its best reference match. This can be useful if the user really needs species or genus classifications but many organisms in the studied species group have similar rRNA sequences, making it hard for the Metaxa2 classifier to classify sequences to the species level.
The Metaxa2 Rarefaction analysis tool (metaxa2_rf) performs a resampling rarefaction analysis (2) based on the output from the Metaxa2 classifier, taking into account also the unclassified portion of rRNAs. The Metaxa2 Uniqueness of Community analyzer (metaxa2_uc), finally, allows analysis of whether the community composition of two or more samples or groups is significantly different. Using resampling of the community data, the null hypothesis that the taxonomic content of two communities is drawn from the same set of taxa (given certain abundances) is tested. All these tools are further described in the manual and the recent paper (1).
The latest version of Metaxa2, including the Metaxa2 Diversity Tools, can be downloaded here.
References
- Bengtsson-Palme J, Thorell K, Wurzbacher C, Sjöling Å, Nilsson RH: Metaxa2 Diversity Tools: Easing microbial community analysis with Metaxa2. Ecological Informatics, 33, 45–50 (2016). doi: 10.1016/j.ecoinf.2016.04.004 [Paper link]
- Gotelli NJ, Colwell RK: Quantifying biodiversity: procedures and pitfalls in the measurement and comparison of species richness. Ecology Letters, 4, 379–391 (2000). doi:10.1046/j.1461-0248.2001.00230.x
- Bengtsson-Palme J, Hartmann M, Eriksson KM, Pal C, Thorell K, Larsson DGJ, Nilsson RH: Metaxa2: Improved Identification and Taxonomic Classification of Small and Large Subunit rRNA in Metagenomic Data. Molecular Ecology Resources (2015). doi: 10.1111/1755-0998.12399 [Paper link]
- Caporaso JG, Kuczynski J, Stombaugh J et al.: QIIME allows analysis of high-throughput community sequencing data. Nature Methods, 7, 335–336 (2010).
- Schloss PD, Westcott SL, Ryabin T et al.: Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Applied and Environmental Microbiology, 75, 7537–7541 (2009).
New features in Metaxa2 Diversity Tools
Metaxa2 has been updated again today to version 2.1.3. This update adds a few features to the Metaxa2 Diversity Tools (metaxa2_uc and metaxa2_rf). The core Metaxa2 programs remain the same as for the previous Metaxa2 versions. The new features were suggested as part of the review process of a Metaxa2-related manuscript, and we thank the anonymous reviewers for their great suggestions!
New features and bug fixes in this update:
- Added the Chao1, iChao1 and ACE estimators in addition to the original species abundance (“Bengtsson-Palme”) model in metaxa2_rf
- Added the Raup-Crick dissimilarity method to the metaxa2_uc tool
- Added a warning message when data is highly skewed for metaxa2_uc
- Improved robustness of the ‘model’ mode of metaxa2_uc for highly skewed sample groups
- Fixed a bug causing miscalculation of Euclidean distances on binary data in metaxa2_uc
The updated version of Metaxa2 can be downloaded here.
Happy barcoding!
TriMetAss updated to version 1.2
TriMetAss has today been updated to version 1.2. The new version addresses a number of minor issues, some of which I thought was fixed with the previous version. The update can be found here.
The main problem with the previous version of TriMetAss was that the Trinity developers had changed many options in the Trinity software, which rendered more recent versions of Trinity incompatible with TriMetAss. TriMetAss was not the only external software using Trinity that was affected by these changes. As far as my testing goes, these incompatibilities should now be fixed, by improved Trinity version determination in TriMetAss. This is still not a guarantee for future changes though, so just to make sure, use one of the Trinity versions tested with TriMetAss (versions v2.1.1 or trinityrnaseq_r2013_08_14).
This time I would like to thank Artemis Louyakis at the Univesity of Florida and Tatsuya Unno at the Jeju National University (Korea) for their input on TriMetAss.
An update to the Metaxa2 Diversity Tools
I have today uploaded an updated version of Metaxa2 (version 2.1.2). This update primarily improves the memory performance of the Metaxa2 Diversity Tools. The core Metaxa2 programs remain the same as for the previous Metaxa2 versions.
New features and bug fixes in this update:
- Dramatically improved memory performance of metaxa2_uc
- Added the
'min'option to the-sflag in metaxa2_uc, which will cause the program to sample the number of entries present in the smallest sample from each sample - Fixes a bug that disregarded the level specified by the
-loption in metaxa2_si - Minor updates and improvements on the manual
The updated version of Metaxa2 can be downloaded here.
Happy barcoding!
FARAO – The Flexible All-Round Annotation Organizer
A problem with annotating contigs from genomic and metagenomic projects is that there are few tools that allow the visualization of the annotated features, particularly if those features come from different sources. To alleviate this problem, I have (with assistance from Rickard Hammarén and Chandan Pal) over the last years developed a new annotation and read coverage visualization package – FARAO – which we today introduce to the public. FARAO has been used to produce the basis for the the contig annotation figures in my paper on the polluted Indian lake. Storing and visualizing annotation and coverage information in FARAO has a number of advantages. FARAO is able to:
- Integrate annotation and coverage information for the same sequence set, enabling coverage estimates of annotated features
- Scale across millions of sequences and annotated features
- Filter sequences, such that only entries with annotations satisfying certain given criteria will be outputted
- Handle annotation and coverage data produced by a range of different bioinformatics tools
- Handle custom parsers through a flexible interface, allowing for adaption of the software to virtually any bioinformatic tool
- Produce high-quality EPS output
- Integrate with MySQL databases
FARAO is today moved from a private pre-release state to a public beta state. It is still possible that this version contains bug that we have not discovered in our testing. Please send me an e-mail and make us aware of the potential shortcomings of our software if you find any unexpected behavior in this version of FARAO.