Regarding ResearchGate and paper requests
I have recently started to receive requests for full-text versions of my publications on ResearchGate. That’s great, but I have yet to figure out how to send them over, without breaking any agreements. As I am in a somewhat intensive work-period at the moment, please forgive me for not spending time on ResearchGate right now. And if you would like full-text versions of my publications, please send me an e-mail! I’ll be glad to help!
Published paper: ITSx
The paper describing our software tool ITSx has now gone online as an Early View paper on the Methods in Ecology and Evolution website. The software just recently left its beta-status behind, and with the paper out as well, we hope that as many people as possible will find use for the software in barcoding efforts of the ITS region. If you’re not familiar with the software – or its predecessor; the fungal ITS Extractor – here is a brief description of what it does:
ITSx is a Perl-based software tool that extracts the ITS1, 5.8S and ITS2 sequences – as well as full-length ITS sequences – from high-throughput sequencing data sets. To achieve this, we use carefully crafted hidden Markov models (HMMs), computed from large alignments of a total of 20 groups of eukaryotes. Testing has shown that ITSx has close to 100% detection accuracy, and virtually zero false-positive extractions. Additionally, it supports multiple processor cores, and is therefore suitable for running also on very large datasets. It is also able to eliminate non-ITS sequences from a given input dataset.
While ITSx supports extractions of ITS sequences from at least 20 different eukaryotic lineages, we ourselves have considerably less experience with many of the eukaryote groups outside of the fungi. We therefore release ITSx with the intent that the research community will evaluate its performance also in other parts of the eukaryote tree, and if necessary contribute data required to address also those lineages in a thorough way.
The ITSx paper can at the moment be cited as:
Bengtsson-Palme, J., Ryberg, M., Hartmann, M., Branco, S., Wang, Z., Godhe, A., De Wit, P., Sánchez-García, M., Ebersberger, I., de Sousa, F., Amend, A. S., Jumpponen, A., Unterseher, M., Kristiansson, E., Abarenkov, K., Bertrand, Y. J. K., Sanli, K., Eriksson, K. M., Vik, U., Veldre, V., Nilsson, R. H. (2013), Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods in Ecology and Evolution. doi: 10.1111/2041-210X.12073
Metaxa and HMMER 3.1b
As you might be aware, a new version of HMMER is out since late May. You might wonder how Metaxa (relying on HMMER3) will work if you update to the new version of HMMER, and I have finally got around to test it! The answer, according to my somewhat limited testing, is that Metaxa 1.1.2 seems to be working fine with HMMER 3.1.
You might need to go into the database directory (“metaxa_db”; should be located in the same directory as the Metaxa binaries), and remove all the files ending with suffixes .h3f .h3i .h3m and .h3p inside the “HMMs” directory. On most installation, this should not be necessary. Myself, I just plugged HMMER 3.1 in and started Metaxa, but if you get error messages complaining that “Error: bad format, binary auxfiles,
binary auxfiles are in an outdated HMMER format (3/b); please hmmpress your HMM file again”, then you should try removing the files and re-running Metaxa. This might especially be a problem on older Metaxa versions. [Update: Note that this fix will likely not work with ITSx!]
Bear in mind that I have not run thorough testing on Metaxa and HMMER 3.1, and probably won’t for the 1.1.2 version, since there’s a 2.0 version waiting just around the corner…
Additionally, if you experience problems with Megraft, you should try the same fix as for Metaxa, but with the Megraft database directory instead. Regarding ITSx, a minor update will be released very soon, which also will address HMMER 3.1b compatibility. [Update: See this post for how to work around HMMER 3.1 problems with ITSx.]
Happy barcoding everyone!
I have joined ResearchGate
On a side note, I just joined Research Gate (my profile). I’ve noted that it generates kind of the same kind of belonging-to-a-group feeling that registering on Facebook did way back, when co-author after co-author starts following you. Still, I haven’t figured out exactly what to use it for yet; it certainly seem more useful than academia.edu, with abilities to ask questions etc., but is anyone of you really using ResearchGate for this? Or is it rather just another showcasing window for researchers (much like my Publications page)? Please feel free do add your opinions as comments to this post!
Server upgrades
I’ve been informed by my web service provider that there will potentially be downtime of this site on the 13th of February (Wednesday this week), due to a server upgrade. I hope this will cause as little trouble as possible (both for you and for me).
Metagenomics and the Hype Cycle
I was creating the diagram below an upcoming presentation, and I realized that the exponential growth in published metagenomics papers might be coming to an end. Interestingly enough the small drop in pace the recent years (701 -> 983 -> 1148) reminds me of the Hype Cycle, where we would (if my projection holds) have reached the “Peak of Inflated Expectations”, which means that we will see a rapid drop in the number of metagenomics publications in the next few years, as the field moves on.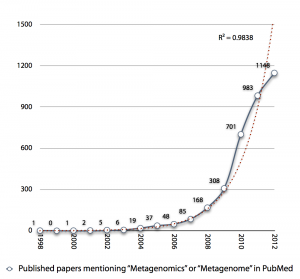
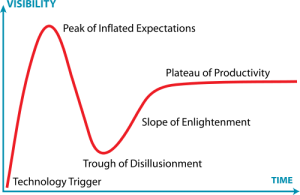 The thought is interesting, but it seems a little bit early to draw any conclusions from the number of publications, yet. It is still kind of strange to note, though, that more than 20% of metagenomics publications (740/3547) are review papers. Come on, let’s do some science first and then review it… Anyway, it’ll be interesting to see what 2013 has in store for us.
The thought is interesting, but it seems a little bit early to draw any conclusions from the number of publications, yet. It is still kind of strange to note, though, that more than 20% of metagenomics publications (740/3547) are review papers. Come on, let’s do some science first and then review it… Anyway, it’ll be interesting to see what 2013 has in store for us.
Introducing the PETKit
You know the feeling when your assembler supports paired-end sequences, but your FASTQ quality filterer doesn’t care about what pairs that belong together? Meaning that you end up with a mess of sequences that you have to script together in some way. Gosh, that feeling is way too common. It is for situations like that I have put together the Paired-End ToolKit (PETKit), a collection of FASTQ/FASTA sequence handling programs written in Perl. Currently the toolkit contains three command-line tools that does sequence conversion, quality filtering, and ORF prediction, all adapted for paired-end sequences specifically. You can read more about the programs, which are released as open source software, on the PETKit page. At the moment they lack proper documentation, but running the software with the “–help” option should bring up a useful set of options for each tool. This is still considered beta-software, so any bug reports, and especially suggestions, are welcome.
Also, if you have an idea of another problem that is unsolved or badly executed for paired-end sequences, let me know, and I will see if I can implement it in PETKit.
Looking for a job?
The Core Facilites at Sahlgrenska are looking for a skilled bioinformatician that can support research projects employing the Core Facilites’ services. The employee will e.g. deal with setting up analysis pipelines for next generation sequencing data. They (of course) want an experienced bioinformatician, who also knows programming (Java, C and/or C++, and scripting languages such as Perl or Python). It is also preferable if the applicant knows how to set up secure systems and manage work with the Unix/Linux terminal. More on the position can be found at GU’s web site. The application time closes on the 17th of September.
Improving Swedish research – is there a need for a research elite?
I know that this is not supposed to be a political page, but writing this up, I realized that there is no way I can keep my political views entirely out of this post. So just a quick warning, the following text contains political opinions and is a reflection of my views and believes rather than well supported facts.
So, Swedish minister for education Jan Björklund has announced the government’s plan to spend 3 billion SEK (~350 million EUR, ~450 million USD) on “elite” researchers over the next ten years. One main reason to do so is to strengthen Swedish research in competition with American universities, and to be able to recruit top researchers from other countries to Sweden. While I welcome the prospect of more money to research, I have to say I am very skeptical about the nature of how this money is distributed. First of all, giving more money to the researchers that have already succeeded (I guess this is how you would define elite researchers – if someone has a better idea, please tell both me and Jan Björklund), is not going to generate more innovative research – just more of the same (or similar) things to what these researchers already do. If the government is serious about that Swedish research has a lower-than-expected output (which is a questionable statement in itself), the best way of increasing that output would be to give more researchers the opportunity to put their ideas into action. Second, a huge problem for research in Sweden is that a lot of the scientists’ time is spent on doing other stuff – writing grant applications, administering courses, filling in forms etc. Therefore, one way of improving research would be to put more money into funding at the university administration level, so that researchers actually have time to do what they are supposed to do. I will now provide my own four-point program for how I think that Sweden should move forward to improve the output of science.
1. Researchers need more time
My first point is that researchers need more time to do what they are supposed to do – science. This means that they cannot be expected to apply for money from six different research foundations every year, just to receive a very small amount of money that will keep them from getting thrown out for another 8 months. The short-term contracts that are currently the norm in Sweden create a system where way too much time is spent on writing grant applications – the majority of which will not succeed. In addition, researchers are often expected to be their own secretary, as well as organizing courses (not only lecturing). To solve this we need:
- Longer contracts for scientists. A grant should be large enough to secure five years of salary, plus equipment costs. This allows for some time to actually get the science done, not just the time to write the next application.
- Grants that come with a guaranteed five-year extension of grants to projects that have fulfilled their goals in the first five years. This further secures longevity of researchers and their projects. Also, this allows for universities to actually employ scientists instead of the current system which is all about trying to work around the employment rules.
- More money to university administration. It is simple more cost efficient to have a secretary handling non-science related stuff in the department or group, as well as economic people handling the economy. The current system expects every researcher to be a jack of all trades – which efficiently reduces one to a master of none. More money to administration means more time spent on research.
2. Broad funding creates a foundation for success
Another problem is that if only a few projects are funded repeatedly, the success of Swedish research is very much bound to the success of these projects. While large-scale and high-cost projects are definitely needed, there is also a need to invest in a variety of projects. Many applied ideas have originated from very non-applied research, and the applied research need fundamental research to be done to be able to move forward. However, in the shortsighted governmental view of science, the output has to be almost immediate, which means that applied projects are much more likely to be funded. Thus, projects that could do fundamental discoveries, but are more complicated and take longer time will be down-prioritized by both researchers and universities. To further make situation worse, Björklund et al. have promised more money to universities that cut out non-productive research, with almost guarantees that any projects with a ten-year timeframe will not even be started.
If we are serious about making Swedish research successful, we need to do exactly the opposite. Fund a lot of different projects, both applied and fundamental, regardless of their short-term value. Because the ideas that are most likely to produce short-term results are probably also the ones that are the least innovative in the long-term. Consequently, we need to:
- Spend research funding on a variety of projects, both of fundamental and applied nature.
- Secure funding for “crazy” projects that span long periods of time, at least five to ten years.
3. If we don’t dare to fail, we will not have a chance to win
Finally, research funding must become better at taking risks. If we only bet our money on the most successful researchers, there is absolutely no chance for young scientists to get funded, unless of course they have been picked up by one of the right supervisors. This means that the same ideas get disseminated through the system over and over again, at the expense of more innovative ideas that could pop up in groups with less money to realize them. If these untested ideas in smaller groups get funded, some of them might undoubtedly fail to produce research of high societal value. But some of them will likely develop entirely new ideas, which in the long term might be much more fruitful than throwing money on the same groups over and over again. Suggestions:
- Spend research funding broadly and with an active risk-gain management strategy.
- Allow for fundamental research to investigate completely new concepts – even if they are previously untested, and regardless (or less dependent on) previous research output.
- Invest in infrastructure for innovative research – and do so fast. For example, the money spent on the sequencing facilities at Sci Life Lab in Stockholm is an excellent example of an infrastructure investment that gains a lot of researchers at different universities access to high-throughput sequencing, without each university having to invest in expensive sequencing platforms themselves. More such centers would both spur collaboration and allow for faster adoption of new technologies.
4. Competing with what we are best at
A mistake that is often done when trying to compete with those that are best in the class is to try to compete by doing the same things as the best players do. This makes it extremely hard to win a game against exactly those players, as they are likely more experienced, have more resources, and already has the attention to get the resources we compete for. Instead, one could try to play the Wayne Gretzky trick: to try to skate where the puck is heading, instead of where it is today. Another approach would be to invent a new arena for the puck to land in, where you have better control over the settings than your competitors (slightly similar to what Apple did when the iPod was released, and Microsoft couldn’t use Windows to leverage their mp3-player Zune).
For Sweden, this would mean that we should not throw some bucks at the best players at our universities and hope that they will be happy with this (comparably small) amount of money. Instead, we should give them circumstances to work under that are much better or appealing from other standpoints. This could be better job security, longer contracts, less administrative work, securer grants, more freedom to decide over ones time, and larger possibilities to combine work and family. Simply creating a better, securer and nicer environment to work in. However, Björklund’s suggestions go the very opposite way: researchers should compete to be part of the elite community, and if your not in that group, you’d get thrown out. Therefore, I suggest (with the risk of repeating myself) that we should compete by:
- Offering longer contracts and grants for scientists.
- Giving scientists opportunities to combine work and family life.
- Embracing all kinds of science, both fundamental and applied, both short-term and long-term.
- Allowing researchers to take risks, even if they fail.
- Giving universities enough funding to let scientists do the science and administrative personal do the administration.
- Funding large-scale collaborative infrastructure investments.
- Thinking of how to create an environment that is appealing for scientists, not only from an economic perspective.
A note on other important aspects of funding
Finally, I have now been focusing a lot on width as opposed to directed funding to an elite research squad. It is, however, apparent that we also need to allocate funding to bring in more women to the top positions in the academy. Likely, a system which favors elite groups will also favor male researchers, judging from how the Swedish Foundation for Strategic Research picks their bets for the future. Also, it is important that young researchers without strong track records gets funded, otherwise a lot of new and interesting ideas risk to be lost.
In the fourth point of my proposal, I suggest that Sweden should compete at what Sweden is good at, that is to view researchers as human beings, which are most likely to succeed in an environment where they can develop their ideas in a free and secure way. For me, it is surprising that a minister of education representing a liberal party wants to excess such control over what is good and bad research. Putting up a working social security system around science seems much more logical than throwing money at those who already have. Apparently I have forgotten that our current government is not interested in having a working social security system – their interest seem to lie in deconstructing the very same structures.
ISME14 begins today
I am on my way to Copenhagen for the ISME14 conference that begins today. I’m myself quite excited about this event, and will present three posters (two as first author), and give a short talk on antibiotic resistance gene identification and metagenomics. My talk will be in the Bioinformatics in Microbial Ecology session on Thursday afternoon (at 13.30).
 If you’d like to talk about Metaxa and Megraft, I will present an SSU-oriented poster in the Monday afternoon poster section (board number 267A). My antibiotic resistance gene poster will be presented on Thursday afternoon (board number 002A), and I really encourage everyone interested in metagenomics (especially metagenomic assembly) to come talk to me then! Finally, I am also partially responsible for a poster on periphyton metagenomics with Martin Eriksson as its main author. This poster is also presented on Monday, in the Microbial Dispersion and Biogeography session (board number 021A).
If you’d like to talk about Metaxa and Megraft, I will present an SSU-oriented poster in the Monday afternoon poster section (board number 267A). My antibiotic resistance gene poster will be presented on Thursday afternoon (board number 002A), and I really encourage everyone interested in metagenomics (especially metagenomic assembly) to come talk to me then! Finally, I am also partially responsible for a poster on periphyton metagenomics with Martin Eriksson as its main author. This poster is also presented on Monday, in the Microbial Dispersion and Biogeography session (board number 021A).
I hope to be able to make another post later tonight on what are the “essential” sessions for me on this conference. Hope to see you there soon!